Dissertation Domik 2016.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Structure-Based Virtual Screening of Hypothetical Inhibitors of the Enzyme Longiborneol Synthase—A Potential Target to Reduce Fusarium Head Blight Disease
J Mol Model (2016) 22: 163 DOI 10.1007/s00894-016-3021-1 ORIGINAL PAPER Structure-based virtual screening of hypothetical inhibitors of the enzyme longiborneol synthase—a potential target to reduce Fusarium head blight disease E. Bresso1 & V. L ero ux 2 & M. Urban3 & K. E. Hammond-Kosack3 & B. Maigret2 & N. F. Martins1 Received: 17 December 2015 /Accepted: 27 May 2016 /Published online: 21 June 2016 # Springer-Verlag Berlin Heidelberg 2016 Abstract Fusarium head blight (FHB) is one of the most compounds from a library of 15,000 drug-like compounds. destructive diseases of wheat and other cereals worldwide. These putative inhibitors of longiborneol synthase provide a During infection, the Fusarium fungi produce mycotoxins that sound starting point for further studies involving molecular represent a high risk to human and animal health. Developing modeling coupled to biochemical experiments. This process small-molecule inhibitors to specifically reduce mycotoxin could eventually lead to the development of novel approaches levels would be highly beneficial since current treatments to reduce mycotoxin contamination in harvested grain. unspecifically target the Fusarium pathogen. Culmorin pos- sesses a well-known important synergistically virulence role Keywords Fusarium mycotoxins . Culmorin . Inhibitors . among mycotoxins, and longiborneol synthase appears to be a Homology modeling . Molecular dynamics . Ensemble key enzyme for its synthesis, thus making longiborneol syn- docking thase a particularly interesting target. This study aims to dis- cover potent and less toxic agrochemicals against FHB. These compounds would hamper culmorin synthesis by inhibiting Introduction longiborneol synthase. In order to select starting molecules for further investigation, we have conducted a structure- Fusarium head blight (FHB), caused by Fusarium based virtual screening investigation. -

Characterization of a Cytochrome P450 Monooxygenase Gene Involved in the Biosynthesis of Geosmin in Penicillium Expansum
Open Archive TOULOUSE Archive Ouverte (OATAO) OATAO is an open access repository that collects the work of Toulouse researchers and makes it freely available over the web where possible. This is an author-deposited version published in : http://oatao.univ-toulouse.fr/ Eprints ID : 10812 To link to this article : DOI : 10.5897/AJMR11.1361 URL : http://academicjournals.org/journal/AJMR/article- abstract/841ECBA21771 To cite this version : Siddique, Muhammad Hussnain and Liboz, Thierry and Bacha, Nafees and Puel, Olivier and Mathieu, Florence and Lebrihi, Ahmed Characterization of a cytochrome P450 monooxygenase gene involved in the biosynthesis of geosmin in Penicillium expansum. (2012) African Journal of Microbiology Research, vol. 6 (n° 19). pp. 4122-4127. ISSN 1996-0808 Any correspondance concerning this service should be sent to the repository administrator: [email protected] Characterization of a cytochrome P450 monooxygenase gene involved in the biosynthesis of geosmin in Penicillium expansum Muhammad Hussnain Siddique1,2, Thierry Liboz1,2, Nafees Bacha3, Olivier Puel4, Florence Mathieu1,2 and Ahmed Lebrihi1,2,5* 1Université de Toulouse, INPT-UPS, Laboratoire de Génie Chimique, avenue de l’Agrobiopole, 31326 Castanet-Tolosan Cedex, France. 2Le Centre national de la recherche scientifique (CNRS), Laboratoire de Génie Chimique, 31030 Toulouse, France. 3Centre of Biotechnology and Microbiology, University of Peshawar, Pakistan. 4Institut National de la Recherche Agronomique (INRA), Laboratoire de Pharmacologie Toxicologie, 31931 Toulouse, France. 5Université Moulay Ismail, Marjane 2, BP 298, Meknes, Morocco. Geosmin is a terpenoid, an earthy-smelling substance associated with off-flavors in water and wine. The biosynthesis of geosmin is well characterized in bacteria, but little is known about its production in eukaryotes, especially in filamentous fungi. -

Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000725805Cyc: Streptomyces xanthophaeus Cellular Overview Connections between pathways are omitted for legibility. -

Production of Plant-Associated Volatiles by Select Model and Industrially Important Streptomyces Spp
microorganisms Article Production of Plant-Associated Volatiles by Select Model and Industrially Important Streptomyces spp. 1, 2, 3 1 Zhenlong Cheng y, Sean McCann y, Nicoletta Faraone , Jody-Ann Clarke , E. Abbie Hudson 2, Kevin Cloonan 2, N. Kirk Hillier 2,* and Kapil Tahlan 1,* 1 Department of Biology, Memorial University of Newfoundland, St. John’s, NL A1B 3X9, Canada; [email protected] (Z.C.); [email protected] (J.-A.C.) 2 Department of Biology, Acadia University, Wolfville, NS B4P 2R6, Canada; [email protected] (S.M.); [email protected] (E.A.H.); [email protected] (K.C.) 3 Department of Chemistry, Acadia University, Wolfville, NS B4P 2R6, Canada; [email protected] * Correspondence: [email protected] (N.K.H.); [email protected] (K.T.) These authors contributed equally. y Received: 13 October 2020; Accepted: 9 November 2020; Published: 11 November 2020 Abstract: The Streptomyces produce a great diversity of specialized metabolites, including highly volatile compounds with potential biological activities. Volatile organic compounds (VOCs) produced by nine Streptomyces spp., some of which are of industrial importance, were collected and identified using gas chromatography–mass spectrometry (GC-MS). Biosynthetic gene clusters (BGCs) present in the genomes of the respective Streptomyces spp. were also predicted to match them with the VOCs detected. Overall, 33 specific VOCs were identified, of which the production of 16 has not been previously reported in the Streptomyces. Among chemical classes, the most abundant VOCs were terpenes, which is consistent with predicted biosynthetic capabilities. In addition, 27 of the identified VOCs were plant-associated, demonstrating that some Streptomyces spp. -

Discovery of Cryptic Compounds from Streptomyces Lavendulae FRI-5 Using an Engineered Microbial Host (異種発現系の活用による放線菌休眠化合物の覚醒と同定)
Discovery of cryptic compounds from Streptomyces Title lavendulae FRI-5 using an engineered microbial host Author(s) Pait, Ivy Grace Citation Issue Date Text Version ETD URL https://doi.org/10.18910/69537 DOI 10.18910/69537 rights Note Osaka University Knowledge Archive : OUKA https://ir.library.osaka-u.ac.jp/ Osaka University Doctoral Dissertation Discovery of cryptic compounds from Streptomyces lavendulae FRI-5 using an engineered microbial host (異種発現系の活用による放線菌休眠化合物の覚醒と同定) Ivy Grace Umadhay Pait January 2018 Division of Advanced Science and Biotechnology Graduate School of Engineering, Osaka University Contents Chapter 1 General Introduction 1.1 The role of microbial natural products in drug discovery 1 1.2 Bioactivities and biosynthesis of polyketides and nonribosomal peptides 3 1.2.1 The assembly-line enzymology of polyketides 3 1.2.2 The biosynthetic logic of nonribosomal peptide compounds 7 1.3 Streptomyces, the proven resource for therapeutics 9 1.3.1 Life cycle of Streptomyces 10 1.3.2 Regulation of secondary metabolite production in Streptomyces 11 1.4 Genome mining and reviving interest in microbial secondary metabolites 14 1.4.1 Decline in natural product research 14 1.4.2 Bacterial genome mining for new natural products 16 1.4.3 Streptomyces genome as the richest bacterial resource for cryptic BGCs 17 1.4.4 Approaches for triggering the production of cryptic metabolites 19 1.4.4.1 Altering chemical and physical conditions 19 1.4.4.2 Genetic modification/Molecular approaches 21 1.5 Heterologous expression in a genome-minimized -

Physarum Polycephalum) by SPME
Analysis of the volatiles in the headspace above the plasmodium and sporangia of the slime mould (Physarum polycephalum) by SPME- GCMS Huda al Kateb1 and Ben de Lacy Costello1 1Institute for biosensing technology, University of the West of England, Bristol, BS161QY, UK E-mail: [email protected] Abstract Solid phase micro-extraction (SPME) coupled with Gas Chromatography Mass Spectrometry (GC-MS) was used to extract and analyse the volatiles in the headspace above the plasmodial and sporulating stages of the slime mould Physarum Polycephalum. In total 115 compounds were identified from across a broad range of chemical classes. Although more (87) volatile organic compounds (VOCs) were identified when using a higher incubation temperature of 75oC, a large number of compounds (79) were still identified at the lower extraction temperature of 30oC and where the plasmodial stage was living. Far fewer compounds were extracted after sporulation at the two extraction temperatures. There were some marked differences between the VOCs identified in the plasmodial stage and after sporulation. In particular the nitrogen containing compounds acetonitrile, pyrrole, 2, 5-dimethyl-pyrazine and trimethyl pyrazine seemed to be associated with the sporulating stage. There were many compounds associated predominantly with the plasmodial stage including a number of furans and alkanes. Interestingly, a number of known fungal metabolites were identified including 1-octen-3- ol, 3-octanone, 1-octen-3-one, 3-octanol. In addition known metabolites of cyanobacteria and actinobacteria in particular geosmin was identified in the headspace. Volatile metabolites that had previously been identified as having a positive chemotactic response to the plasmodial stage of P. -

Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_900114035Cyc: Amycolatopsis sacchari DSM 44468 Cellular Overview Connections between pathways are omitted for legibility. -
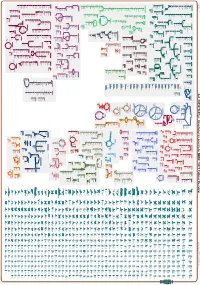
Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000367365Cyc: Streptomyces prunicolor NBRC 13075 Cellular Overview Connections between pathways are omitted for legibility. -

Biokatalytische Diversität Der Terpenbildung in Pflanzen Und Bakterien”
Biokatalytische Diversität der Terpenbildung in Pflanzen und Bakterien Dissertation zur Erlangung des Grades eines Doktors der Naturwissenschaften der Fakultät für Chemie und Biochemie an der Graduate School for Chemistry and Biochemistry der Ruhr-Universität Bochum angefertigt in der Nachwuchsgruppe für Mikrobielle Biotechnologie vorgelegt von Octavia Natascha Kracht aus Unna Bochum Juli 2017 Erstgutachter: Prof. Dr. Robert Kourist Zweitgutachter: Jun.-Prof. Dr. Simon Ebbinghaus Diese Arbeit wurde in der Zeit von Mai 2014 bis Juli 2017 unter der Leitung von Jun.- Prof. Dr. Robert Kourist in der Nachwuchsgruppe für Mikrobielle Biotechnologie an der Ruhr-Universität Bochum durchgeführt. 2 Danksagung An dieser Stelle möchte ich mich bei den Personen bedanken, die mich während der Anfertigung dieser Arbeit immer unterstützt und damit einen Großteil zum Gelingen dieses Projektes beigetragen haben. Mein größter Dank gilt meinem Doktorvater Prof. Dr. Robert Kourist für die interessante Themenstellung und die Möglichkeit der Anfertigung dieser Dissertation in seiner Arbeitsgruppe. Vielen Dank für das Vertrauen, dass du mir entgegengebracht hast und deine sowohl fachliche als auch persönliche Unterstützung während meiner gesamten Promotion. Auch auf langen Durststrecken hast du immer an unser Projekt geglaubt und mich stets motiviert. Vielen Dank für deine ständige Gesprächsbereitschaft, die konstruktiven Beiträge und vor allem die Ermöglichung meines Auslandsaufenthaltes in Kanada. Ich werde meine Zeit in deiner Gruppe immer in guter Erinnerung behalten. Ich möchte mich außerdem ganz herzlich bei Jun.-Prof. Dr. Simon Ebbinghaus für die freundliche Übernahme des Koreferates bedanken. Einen ganz großen Dank möchte ich unseren Gärtnern Andreas Aufermann und Martin Pullack (LS Pflanzenphysiologie, RUB) ausprechen. Ohne euch wäre die Anfertigung dieser Arbeit gar nicht erst möglich gewesen! Egal ob Weiße Fliege, Läuse oder Behandlung mit Methanol, ihr habt nie aufgegeben und euch immer etwas Neues einfallen lassen, um unsere Pflanzen zu erhalten. -

Hydroxylation of 1-Deoxypentalenic Acid Mediated by CYP105D7 (SAV 7469) of Streptomyces Avermitilis
The Journal of Antibiotics (2011) 64, 65–71 & 2011 Japan Antibiotics Research Association All rights reserved 0021-8820/11 $32.00 www.nature.com/ja ORIGINAL ARTICLE Pentalenic acid is a shunt metabolite in the biosynthesis of the pentalenolactone family of metabolites: hydroxylation of 1-deoxypentalenic acid mediated by CYP105D7 (SAV_7469) of Streptomyces avermitilis Satoshi Takamatsu1,4, Lian-Hua Xu2,4, Shinya Fushinobu2, Hirofumi Shoun2, Mamoru Komatsu1, David E Cane3 and Haruo Ikeda1 Pentalenic acid (1) has been isolated from many Streptomyces sp. as a co-metabolite of the sesquiterpenoid antibiotic pentalenolactone and related natural products. We have previously reported the identification of a 13.4-kb gene cluster in the genome of Streptomyces avermitilis implicated in the biosynthesis of the pentalenolactone family of metabolites consisting of 13 open reading frames. Detailed molecular genetic and biochemical studies have revealed that at least seven genes are involved in the biosynthesis of the newly discovered metabolites, neopentalenoketolactone, but no gene specifically dedicated to the formation of pentalenic acid (1) was evident in the same gene cluster. The wild-type strain of S. avermitilis, as well as its derivatives, mainly produce pentalenic acid (1), together with neopentalenoketolactone (9). Disruption of the sav7469 gene encoding a cytochrome P450 (CYP105D7), members of which class are associated with the hydroxylation of many structurally different compounds, abolished the production of pentalenic acid (1). The sav7469-deletion mutant derived from SUKA11 carrying pKU462Hptl-clusterDptlH accumulated 1-deoxypentalenic acid (5), but not pentalenic acid (1). Reintroduction of an extra-copy of the sav7469 gene to SUKA11 Dsav7469 carrying pKU462Hptl-clusterDptlH restored the production of pentalenic acid (1). -

Geosmin Biosynthesis in Streptomyces Avermitilis. Molecular Cloning, Expression, and Mechanistic Study of the Germacradienol/Geosmin Synthase David E
J. Antibiot. 59(8): 471–479, 2006 THE JOURNAL OF ORIGINAL ARTICLE [_ ANTIBIOTICSJ Geosmin Biosynthesis in Streptomyces avermitilis. Molecular Cloning, Expression, and Mechanistic Study of the Germacradienol/Geosmin Synthase David E. Cane, Xiaofei He, Seiji Kobayashi, Satoshi O¯ mura, Haruo Ikeda This paper is submitted in honor of the late Professor Kenneth L. Rinehart, a pioneer in the study of natural products and their biosynthesis. Received: May 30, 2006 / Accepted: August 3, 2006 © Japan Antibiotics Research Association Abstract Geosmin (1) is responsible for the Keywords sesquiterpene, biosynthesis, geosmin, characteristic odor of moist soil. The Gram-positive soil Streptomyces avermitilis, Streptomyces coelicolor bacterium Streptomyces avermitilis produces geosmin (1) as well as its precursor germacradienol (3). The S. avermitilis gene SAV2163 (geoA) is extremely similar to Introduction the S. coelicolor A3(2) SCO6073 gene that encodes a germacradienol/geosmin synthase. S. avermitilis mutants Geosmin (1) is a well-known, odoriferous metabolite with a deleted geoA were unable to produce either produced by a wide variety of microorganisms, including germacradienol (3) or geosmin (1). Biosynthesis of both Streptomyces, cyanobacteria, myxobacteria, and various compounds was restored by introducing an intact geoA fungi, as well as by higher plants such as liverworts and gene into the mutants. Incubation of recombinant GeoA, sugar beets [1ϳ7]. Geosmin, with an especially low encoded by the SAV2163 gene of S. avermitilis, with detection threshold of 10ϳ100 parts per trillion, is farnesyl diphosphate (2) in the presence of Mg2ϩ gave a responsible for the characteristic odor of freshly turned mixture of (4S,7R)-germacra-1(10)E,5E-diene-11-ol (3) earth and is associated with unpleasant off-flavors in water, (66%), (7S)-germacrene D (4) (24%), geosmin (1) (8%), wine, and fish [8, 9]. -

Generated by SRI International Pathway Tools Version 25.0 on Thu
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_900105695Cyc: Streptomyces melanosporofaciens DSM 40318 Cellular Overview Connections between pathways are omitted for legibility.