Physarum Polycephalum) by SPME
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Structure-Based Virtual Screening of Hypothetical Inhibitors of the Enzyme Longiborneol Synthase—A Potential Target to Reduce Fusarium Head Blight Disease
J Mol Model (2016) 22: 163 DOI 10.1007/s00894-016-3021-1 ORIGINAL PAPER Structure-based virtual screening of hypothetical inhibitors of the enzyme longiborneol synthase—a potential target to reduce Fusarium head blight disease E. Bresso1 & V. L ero ux 2 & M. Urban3 & K. E. Hammond-Kosack3 & B. Maigret2 & N. F. Martins1 Received: 17 December 2015 /Accepted: 27 May 2016 /Published online: 21 June 2016 # Springer-Verlag Berlin Heidelberg 2016 Abstract Fusarium head blight (FHB) is one of the most compounds from a library of 15,000 drug-like compounds. destructive diseases of wheat and other cereals worldwide. These putative inhibitors of longiborneol synthase provide a During infection, the Fusarium fungi produce mycotoxins that sound starting point for further studies involving molecular represent a high risk to human and animal health. Developing modeling coupled to biochemical experiments. This process small-molecule inhibitors to specifically reduce mycotoxin could eventually lead to the development of novel approaches levels would be highly beneficial since current treatments to reduce mycotoxin contamination in harvested grain. unspecifically target the Fusarium pathogen. Culmorin pos- sesses a well-known important synergistically virulence role Keywords Fusarium mycotoxins . Culmorin . Inhibitors . among mycotoxins, and longiborneol synthase appears to be a Homology modeling . Molecular dynamics . Ensemble key enzyme for its synthesis, thus making longiborneol syn- docking thase a particularly interesting target. This study aims to dis- cover potent and less toxic agrochemicals against FHB. These compounds would hamper culmorin synthesis by inhibiting Introduction longiborneol synthase. In order to select starting molecules for further investigation, we have conducted a structure- Fusarium head blight (FHB), caused by Fusarium based virtual screening investigation. -

Characterization of a Cytochrome P450 Monooxygenase Gene Involved in the Biosynthesis of Geosmin in Penicillium Expansum
Open Archive TOULOUSE Archive Ouverte (OATAO) OATAO is an open access repository that collects the work of Toulouse researchers and makes it freely available over the web where possible. This is an author-deposited version published in : http://oatao.univ-toulouse.fr/ Eprints ID : 10812 To link to this article : DOI : 10.5897/AJMR11.1361 URL : http://academicjournals.org/journal/AJMR/article- abstract/841ECBA21771 To cite this version : Siddique, Muhammad Hussnain and Liboz, Thierry and Bacha, Nafees and Puel, Olivier and Mathieu, Florence and Lebrihi, Ahmed Characterization of a cytochrome P450 monooxygenase gene involved in the biosynthesis of geosmin in Penicillium expansum. (2012) African Journal of Microbiology Research, vol. 6 (n° 19). pp. 4122-4127. ISSN 1996-0808 Any correspondance concerning this service should be sent to the repository administrator: [email protected] Characterization of a cytochrome P450 monooxygenase gene involved in the biosynthesis of geosmin in Penicillium expansum Muhammad Hussnain Siddique1,2, Thierry Liboz1,2, Nafees Bacha3, Olivier Puel4, Florence Mathieu1,2 and Ahmed Lebrihi1,2,5* 1Université de Toulouse, INPT-UPS, Laboratoire de Génie Chimique, avenue de l’Agrobiopole, 31326 Castanet-Tolosan Cedex, France. 2Le Centre national de la recherche scientifique (CNRS), Laboratoire de Génie Chimique, 31030 Toulouse, France. 3Centre of Biotechnology and Microbiology, University of Peshawar, Pakistan. 4Institut National de la Recherche Agronomique (INRA), Laboratoire de Pharmacologie Toxicologie, 31931 Toulouse, France. 5Université Moulay Ismail, Marjane 2, BP 298, Meknes, Morocco. Geosmin is a terpenoid, an earthy-smelling substance associated with off-flavors in water and wine. The biosynthesis of geosmin is well characterized in bacteria, but little is known about its production in eukaryotes, especially in filamentous fungi. -

Sounds Synthesis with Slime Mould of Physarum Polycephalum
Miranda, Adamatzky, Jones, Journal of Bionic Engineering 8 (2011) 107–113. Sounds Synthesis with Slime Mould of Physarum Polycephalum Eduardo R. Miranda1, Andrew Adamatzky2 and Jeff Jones2 1 Interdisciplinary Centre for Computer Music Research (ICCMR), University of Plymouth, Plymouth, PL4 8AA UK; [email protected] 2 Unconventional Computing Centre, University of the West of England, Bristol, BS16 1QY UK; [email protected] Abstract Physarum polycephalum is a huge single cell with thousands of nuclei, which behaves like a giant amoeba. During its foraging behaviour this plasmodium produces electrical activity corresponding to different physiological states. We developed a method to render sounds from such electrical activity and thus represent spatio-temporal behaviour of slime mould in a form apprehended by humans. We show to control behaviour of slime mould to shape it towards reproduction of required range of sounds. 1 Introduction Our research is concerned with the application of novel computational paradigms implemented on biological substrates in the field of computer music. Computer music has evolved in tandem with the field of Computer Science. Computers have been programmed to produce sounds as early as the beginning of the 1950’s. Nowadays, the computer is ubiquitous in many aspects of music, ranging from software for musical composition and production, to systems for distribution of music on the Internet. Therefore, it is likely that future developments in fields such as Bionic Engineering will have an impact in computer music applications. Research into novel computing paradigms in looking for new algorithms and computing architectures inspired by, or physically implemented on, chemical, biological and physical substrates (Calude et al. -

Biodiversity of Plasmodial Slime Moulds (Myxogastria): Measurement and Interpretation
Protistology 1 (4), 161–178 (2000) Protistology August, 2000 Biodiversity of plasmodial slime moulds (Myxogastria): measurement and interpretation Yuri K. Novozhilova, Martin Schnittlerb, InnaV. Zemlianskaiac and Konstantin A. Fefelovd a V.L.Komarov Botanical Institute of the Russian Academy of Sciences, St. Petersburg, Russia, b Fairmont State College, Fairmont, West Virginia, U.S.A., c Volgograd Medical Academy, Department of Pharmacology and Botany, Volgograd, Russia, d Ural State University, Department of Botany, Yekaterinburg, Russia Summary For myxomycetes the understanding of their diversity and of their ecological function remains underdeveloped. Various problems in recording myxomycetes and analysis of their diversity are discussed by the examples taken from tundra, boreal, and arid areas of Russia and Kazakhstan. Recent advances in inventory of some regions of these areas are summarised. A rapid technique of moist chamber cultures can be used to obtain quantitative estimates of myxomycete species diversity and species abundance. Substrate sampling and species isolation by the moist chamber technique are indispensable for myxomycete inventory, measurement of species richness, and species abundance. General principles for the analysis of myxomycete diversity are discussed. Key words: slime moulds, Mycetozoa, Myxomycetes, biodiversity, ecology, distribu- tion, habitats Introduction decay (Madelin, 1984). The life cycle of myxomycetes includes two trophic stages: uninucleate myxoflagellates General patterns of community structure of terrestrial or amoebae, and a multi-nucleate plasmodium (Fig. 1). macro-organisms (plants, animals, and macrofungi) are The entire plasmodium turns almost all into fruit bodies, well known. Some mathematics methods are used for their called sporocarps (sporangia, aethalia, pseudoaethalia, or studying, from which the most popular are the quantita- plasmodiocarps). -

Culturing Slime Mold
Culturing Slime Mold Live Material Care Guide SCIENTIFIC BIO Background FAX! Plasmodial slime mold (phylum Myxomycota) lives in dark, moist environments such as under the bark of decaying logs, among mulch, or beneath decaying leaves. Slime mold classification is once again changing. They were in Protista due to their amoeboid-like properties. In the past, slime molds were considered a fungus because they produce fruiting bodies and spores used for reproduction. Slime molds are a group notable for its unwillingness to be neatly classified! Frequently bright in color and large in size (up to 30 cm in diameter), plasmodial slime molds consist of many amoeba-like cells, which form a mass of protoplasm called myxomycota. The organisms are capable of very slow, creeping movement by means of cytoplasmic streaming. During the reproductive stage, called pseudoplasmodium, slime molds tend to migrate to a well-lit area, such as the top of a log, where less moisture is present. They form into a slug-like mass and produce reproductive fruiting bodies, which contain spores. Under adverse conditions (lack of food, water, light, warmth, or pH changes), the organism dries out and forms a hardened mass called a sclerotium. These sclerotia may also grow fruiting bodies, but do not release spores into the environ- ment until conditions once again become favorable for growth. Spores are transported by wind, which results in the spreading of slime molds to new areas. Sclerotium (unfavorable conditions) Pseudoplasmodium (favorable conditions) Aggregate (plasmodial stage) Amoeba/spores Reproductive Fruiting Bodies Figure 1. Life Cycle of Slime Mold Culturing/Media Slime mold is typically cultured from sclerotia rather than from spores. -

Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000725805Cyc: Streptomyces xanthophaeus Cellular Overview Connections between pathways are omitted for legibility. -

Physarum Polycephalum - Large Stages by Aggregation of Many Small Amoeboid Cells
Overview Life cycle Physarum polycephalum is the most well- The life cycle of Physarum can be roughly di- known and in the laboratories of cell biologists vided into three phases: plasmodium, fruiting most cultivated representative of the slime body and spores. The large, network-shaped molds (myxomycetes), of which there are plasmodia contain numerous nuclei with a about 900 species. Slime molds combine char- double (= diploid) set of chromosomes, which acteristics of fungi (the formation of fruiting divide synchronously when the cell grows. For bodies) and animals (possession of motile sex growth, the plasmodia need to take up food cells), but are not directly related to either of such as protists, bacteria, fungi, lichens, plant them. Instead, they systematically belong to and animal remains. In the laboratory, the plas- the Amoebozoa, which usually contain tiny, modia can be easily fed with oatmeal. single-celled amoebae. The macroscopically visible life form of Physarum represents a gi- gantic amoeba, i.e. a single cell. This life form, known as plasmodium, contains a large num- ber of nuclei and forms a network of veins Fig. 1: Part of the yellow plasmodium of Physarum (Figs. 1-3). With the help of fluid cell plasma polycephalum with system of veins and migration flowing rhythmically in the veins, the plasmo- front. dium slowly moves. In contrast to this one gi- ant cell, other slime molds such as Dictyoste- lium discoideum (Protist of the Year 2011) form Physarum polycephalum - large stages by aggregation of many small amoeboid cells. The slime mold Fig. 3: Two approximately palm-sized slime molds in their natural habitat, here on the rotting branch of a fallen tree in Grunewald, Berlin. -

Slime Molds: Biology and Diversity
Glime, J. M. 2019. Slime Molds: Biology and Diversity. Chapt. 3-1. In: Glime, J. M. Bryophyte Ecology. Volume 2. Bryological 3-1-1 Interaction. Ebook sponsored by Michigan Technological University and the International Association of Bryologists. Last updated 18 July 2020 and available at <https://digitalcommons.mtu.edu/bryophyte-ecology/>. CHAPTER 3-1 SLIME MOLDS: BIOLOGY AND DIVERSITY TABLE OF CONTENTS What are Slime Molds? ....................................................................................................................................... 3-1-2 Identification Difficulties ...................................................................................................................................... 3-1- Reproduction and Colonization ........................................................................................................................... 3-1-5 General Life Cycle ....................................................................................................................................... 3-1-6 Seasonal Changes ......................................................................................................................................... 3-1-7 Environmental Stimuli ............................................................................................................................... 3-1-13 Light .................................................................................................................................................... 3-1-13 pH and Volatile Substances -

Production of Plant-Associated Volatiles by Select Model and Industrially Important Streptomyces Spp
microorganisms Article Production of Plant-Associated Volatiles by Select Model and Industrially Important Streptomyces spp. 1, 2, 3 1 Zhenlong Cheng y, Sean McCann y, Nicoletta Faraone , Jody-Ann Clarke , E. Abbie Hudson 2, Kevin Cloonan 2, N. Kirk Hillier 2,* and Kapil Tahlan 1,* 1 Department of Biology, Memorial University of Newfoundland, St. John’s, NL A1B 3X9, Canada; [email protected] (Z.C.); [email protected] (J.-A.C.) 2 Department of Biology, Acadia University, Wolfville, NS B4P 2R6, Canada; [email protected] (S.M.); [email protected] (E.A.H.); [email protected] (K.C.) 3 Department of Chemistry, Acadia University, Wolfville, NS B4P 2R6, Canada; [email protected] * Correspondence: [email protected] (N.K.H.); [email protected] (K.T.) These authors contributed equally. y Received: 13 October 2020; Accepted: 9 November 2020; Published: 11 November 2020 Abstract: The Streptomyces produce a great diversity of specialized metabolites, including highly volatile compounds with potential biological activities. Volatile organic compounds (VOCs) produced by nine Streptomyces spp., some of which are of industrial importance, were collected and identified using gas chromatography–mass spectrometry (GC-MS). Biosynthetic gene clusters (BGCs) present in the genomes of the respective Streptomyces spp. were also predicted to match them with the VOCs detected. Overall, 33 specific VOCs were identified, of which the production of 16 has not been previously reported in the Streptomyces. Among chemical classes, the most abundant VOCs were terpenes, which is consistent with predicted biosynthetic capabilities. In addition, 27 of the identified VOCs were plant-associated, demonstrating that some Streptomyces spp. -

Discovery of Cryptic Compounds from Streptomyces Lavendulae FRI-5 Using an Engineered Microbial Host (異種発現系の活用による放線菌休眠化合物の覚醒と同定)
Discovery of cryptic compounds from Streptomyces Title lavendulae FRI-5 using an engineered microbial host Author(s) Pait, Ivy Grace Citation Issue Date Text Version ETD URL https://doi.org/10.18910/69537 DOI 10.18910/69537 rights Note Osaka University Knowledge Archive : OUKA https://ir.library.osaka-u.ac.jp/ Osaka University Doctoral Dissertation Discovery of cryptic compounds from Streptomyces lavendulae FRI-5 using an engineered microbial host (異種発現系の活用による放線菌休眠化合物の覚醒と同定) Ivy Grace Umadhay Pait January 2018 Division of Advanced Science and Biotechnology Graduate School of Engineering, Osaka University Contents Chapter 1 General Introduction 1.1 The role of microbial natural products in drug discovery 1 1.2 Bioactivities and biosynthesis of polyketides and nonribosomal peptides 3 1.2.1 The assembly-line enzymology of polyketides 3 1.2.2 The biosynthetic logic of nonribosomal peptide compounds 7 1.3 Streptomyces, the proven resource for therapeutics 9 1.3.1 Life cycle of Streptomyces 10 1.3.2 Regulation of secondary metabolite production in Streptomyces 11 1.4 Genome mining and reviving interest in microbial secondary metabolites 14 1.4.1 Decline in natural product research 14 1.4.2 Bacterial genome mining for new natural products 16 1.4.3 Streptomyces genome as the richest bacterial resource for cryptic BGCs 17 1.4.4 Approaches for triggering the production of cryptic metabolites 19 1.4.4.1 Altering chemical and physical conditions 19 1.4.4.2 Genetic modification/Molecular approaches 21 1.5 Heterologous expression in a genome-minimized -

Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_900114035Cyc: Amycolatopsis sacchari DSM 44468 Cellular Overview Connections between pathways are omitted for legibility. -
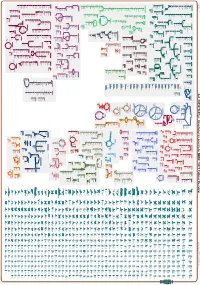
Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000367365Cyc: Streptomyces prunicolor NBRC 13075 Cellular Overview Connections between pathways are omitted for legibility.