The Histone Acetyltransferase Gcne (GCN5) Plays a Central Role in the Regulation of Aspergillus Asexual Development
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Advances in Chitin/Chitosan Characterization and Applications
Advances in Chitin/Chitosan Characterization and Applications Edited by Marguerite Rinaudo and Francisco M. Goycoolea Printed Edition of the Special Issue Published in Polymers www.mdpi.com/journal/polymers Advances in Chitin/Chitosan Characterization and Applications Advances in Chitin/Chitosan Characterization and Applications Special Issue Editors Marguerite Rinaudo Francisco M. Goycoolea MDPI • Basel • Beijing • Wuhan • Barcelona • Belgrade Special Issue Editors Marguerite Rinaudo Francisco M. Goycoolea University of Grenoble Alpes University of Leeds France UK Editorial Office MDPI St. Alban-Anlage 66 4052 Basel, Switzerland This is a reprint of articles from the Special Issue published online in the open access journal Polymers (ISSN 2073-4360) from 2017 to 2018 (available at: https://www.mdpi.com/journal/polymers/ special issues/chitin chitosan) For citation purposes, cite each article independently as indicated on the article page online and as indicated below: LastName, A.A.; LastName, B.B.; LastName, C.C. Article Title. Journal Name Year, Article Number, Page Range. ISBN 978-3-03897-802-2 (Pbk) ISBN 978-3-03897-803-9 (PDF) c 2019 by the authors. Articles in this book are Open Access and distributed under the Creative Commons Attribution (CC BY) license, which allows users to download, copy and build upon published articles, as long as the author and publisher are properly credited, which ensures maximum dissemination and a wider impact of our publications. The book as a whole is distributed by MDPI under the terms and conditions of the Creative Commons license CC BY-NC-ND. Contents About the Special Issue Editors ..................................... ix Preface to ”Advances in Chitin/Chitosan Characterization and Applications” ......... -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000725805Cyc: Streptomyces xanthophaeus Cellular Overview Connections between pathways are omitted for legibility. -

Parasitic Diarrheal Disease: Drug Development and Targets
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Frontiers - Publisher Connector REVIEW published: 27 October 2015 doi: 10.3389/fmicb.2015.01183 Parasitic diarrheal disease: drug development and targets Amir Azam 1*, Mudasir N. Peerzada 1 and Kamal Ahmad 2 1 Medicinal Chemistry Laboratory, Department of Chemistry, Jamia Millia Islamia, New Delhi, India, 2 Centre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia, New Delhi, India Diarrhea is the manifestation of gastrointestinal infection and is one of the major causes of mortality and morbidity specifically among the children of less than 5 years age worldwide. Moreover, in recent years there has been a rise in the number of reports of intestinal infections continuously in the industrialized world. These are largely related to waterborne and food borne outbreaks. These occur by the pathogenesis of both prokaryotic and eukaryotic organisms like bacteria and parasites. The parasitic intestinal infection has remained mostly unexplored and under assessed in terms of therapeutic development. The lack of new drugs and the risk of resistance have led us to carry out this review on drug development for parasitic diarrheal diseases. The major focus has been depicted on commercially available drugs, currently synthesized active heterocyclic compounds and unique drug targets, that are vital for the existence and growth of the parasites and can be further exploited for the search of therapeutically active Edited by: anti-parasitic agents. Anjan Debnath, Keywords: diarrhea, causative parasitic agents, chemotherapy, drug targets, therapeutic developments University of California, San Diego, USA Reviewed by: Ximin Zeng, INTRODUCTION University of Tennessee, USA Sharon Lee Reed, Diarrhea is a symptom of an infection in the intestinal tract, which can be caused by variety of UC San Diego Health Sciences, USA bacterial, viral, and parasitic organisms. -

1995 Aspergillus Bibliography
Fungal Genetics Reports Volume 42 Article 30 1995 Aspergillus Bibliography John Clutterbuck Follow this and additional works at: https://newprairiepress.org/fgr This work is licensed under a Creative Commons Attribution-Share Alike 4.0 License. Recommended Citation Clutterbuck, J. (1995) "1995 Aspergillus Bibliography," Fungal Genetics Reports: Vol. 42, Article 30. https://doi.org/10.4148/1941-4765.1360 This Bibliography is brought to you for free and open access by New Prairie Press. It has been accepted for inclusion in Fungal Genetics Reports by an authorized administrator of New Prairie Press. For more information, please contact [email protected]. 1995 Aspergillus Bibliography Abstract This bibliography attempts to cover genetical and biochemical publications on Aspergillus nidulans and also includes selected references to related species and topics. I would be grateful for publication lists and reprints, especially for papers in books and less readily available periodicals. Entries have been checked as far as possible, but please tell me of any errors. This bibliography is available in Fungal Genetics Reports: https://newprairiepress.org/fgr/vol42/iss1/30 Clutterbuck: 1995 Aspergillus Bibliography Aspergillus Bibliography This bibliography attempts to cover genetical and biochemical publications on Aspergillus nidulans and also includes selected references to related species and topics. I would be grateful for publication lists and reprints, especially for papers in books and less readily available periodicals. Entries have been checked as far as possible, but please tell me of any errors. John Clutterbuck Author and Keyword Index 1. Adams, T.H. 1994 Asexual sporulation in higher fungi, Ch 16 in The Growing Fungus, ed. -

Genome-Wide Investigation of Cellular Functions for Trna Nucleus
Genome-wide Investigation of Cellular Functions for tRNA Nucleus- Cytoplasm Trafficking in the Yeast Saccharomyces cerevisiae DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Hui-Yi Chu Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2012 Dissertation Committee: Anita K. Hopper, Advisor Stephen Osmani Kurt Fredrick Jane Jackman Copyright by Hui-Yi Chu 2012 Abstract In eukaryotic cells tRNAs are transcribed in the nucleus and exported to the cytoplasm for their essential role in protein synthesis. This export event was thought to be unidirectional. Surprisingly, several lines of evidence showed that mature cytoplasmic tRNAs shuttle between nucleus and cytoplasm and their distribution is nutrient-dependent. This newly discovered tRNA retrograde process is conserved from yeast to vertebrates. Although how exactly the tRNA nuclear-cytoplasmic trafficking is regulated is still under investigation, previous studies identified several transporters involved in tRNA subcellular dynamics. At least three members of the β-importin family function in tRNA nuclear-cytoplasmic intracellular movement: (1) Los1 functions in both the tRNA primary export and re-export processes; (2) Mtr10, directly or indirectly, is responsible for the constitutive retrograde import of cytoplasmic tRNA to the nucleus; (3) Msn5 functions solely in the re-export process. In this thesis I focus on the physiological role(s) of the tRNA nuclear retrograde pathway. One possibility is that nuclear accumulation of cytoplasmic tRNA serves to modulate translation of particular transcripts. To test this hypothesis, I compared expression profiles from non-translating mRNAs and polyribosome-bound translating mRNAs collected from msn5Δ and mtr10Δ mutants and wild-type cells, in fed or acute amino acid starvation conditions. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

(12) United States Patent (10) Patent No.: US 8,561,811 B2 Bluchel Et Al
USOO8561811 B2 (12) United States Patent (10) Patent No.: US 8,561,811 B2 Bluchel et al. (45) Date of Patent: Oct. 22, 2013 (54) SUBSTRATE FOR IMMOBILIZING (56) References Cited FUNCTIONAL SUBSTANCES AND METHOD FOR PREPARING THE SAME U.S. PATENT DOCUMENTS 3,952,053 A 4, 1976 Brown, Jr. et al. (71) Applicants: Christian Gert Bluchel, Singapore 4.415,663 A 1 1/1983 Symon et al. (SG); Yanmei Wang, Singapore (SG) 4,576,928 A 3, 1986 Tani et al. 4.915,839 A 4, 1990 Marinaccio et al. (72) Inventors: Christian Gert Bluchel, Singapore 6,946,527 B2 9, 2005 Lemke et al. (SG); Yanmei Wang, Singapore (SG) FOREIGN PATENT DOCUMENTS (73) Assignee: Temasek Polytechnic, Singapore (SG) CN 101596422 A 12/2009 JP 2253813 A 10, 1990 (*) Notice: Subject to any disclaimer, the term of this JP 2258006 A 10, 1990 patent is extended or adjusted under 35 WO O2O2585 A2 1, 2002 U.S.C. 154(b) by 0 days. OTHER PUBLICATIONS (21) Appl. No.: 13/837,254 Inaternational Search Report for PCT/SG2011/000069 mailing date (22) Filed: Mar 15, 2013 of Apr. 12, 2011. Suen, Shing-Yi, et al. “Comparison of Ligand Density and Protein (65) Prior Publication Data Adsorption on Dye Affinity Membranes Using Difference Spacer Arms'. Separation Science and Technology, 35:1 (2000), pp. 69-87. US 2013/0210111A1 Aug. 15, 2013 Related U.S. Application Data Primary Examiner — Chester Barry (62) Division of application No. 13/580,055, filed as (74) Attorney, Agent, or Firm — Cantor Colburn LLP application No. -

Molecular Mechanisms Involved in the Multicellular Growth of Ustilaginomycetes
microorganisms Review Molecular Mechanisms Involved in the Multicellular Growth of Ustilaginomycetes 1,2, , 3, 4 Domingo Martínez-Soto * y, Lucila Ortiz-Castellanos y, Mariana Robledo-Briones and Claudia Geraldine León-Ramírez 3 1 Department of Microbiology and Plant Pathology, University of California, Riverside, CA 92521, USA 2 Tecnológico Nacional de México, Instituto Tecnológico Superior de Los Reyes, Los Reyes 60300, Mexico 3 Departamento de Ingeniería Genética, Unidad Irapuato, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, Irapuato 36821, Mexico; [email protected] (L.O.-C.); [email protected] (C.G.L.-R.) 4 Departamento de Microbiología y Genética, Instituto Hispano-Luso de Investigaciones Agrarias (CIALE), Universidad de Salamanca, 37185 Salamanca, Spain; [email protected] * Correspondence: [email protected] These authors contributed equally. y Received: 7 June 2020; Accepted: 16 July 2020; Published: 18 July 2020 Abstract: Multicellularity is defined as the developmental process by which unicellular organisms became pluricellular during the evolution of complex organisms on Earth. This process requires the convergence of genetic, ecological, and environmental factors. In fungi, mycelial and pseudomycelium growth, snowflake phenotype (where daughter cells remain attached to their stem cells after mitosis), and fruiting bodies have been described as models of multicellular structures. Ustilaginomycetes are Basidiomycota fungi, many of which are pathogens of economically important plant species. These fungi usually grow unicellularly as yeasts (sporidia), but also as simple multicellular forms, such as pseudomycelium, multicellular clusters, or mycelium during plant infection and under different environmental conditions: Nitrogen starvation, nutrient starvation, acid culture media, or with fatty acids as a carbon source. -

POLSKIE TOWARZYSTWO BIOCHEMICZNE Postępy Biochemii
POLSKIE TOWARZYSTWO BIOCHEMICZNE Postępy Biochemii http://rcin.org.pl WSKAZÓWKI DLA AUTORÓW Kwartalnik „Postępy Biochemii” publikuje artykuły monograficzne omawiające wąskie tematy, oraz artykuły przeglądowe referujące szersze zagadnienia z biochemii i nauk pokrewnych. Artykuły pierwszego typu winny w sposób syntetyczny omawiać wybrany temat na podstawie możliwie pełnego piśmiennictwa z kilku ostatnich lat, a artykuły drugiego typu na podstawie piśmiennictwa z ostatnich dwu lat. Objętość takich artykułów nie powinna przekraczać 25 stron maszynopisu (nie licząc ilustracji i piśmiennictwa). Kwartalnik publikuje także artykuły typu minireviews, do 10 stron maszynopisu, z dziedziny zainteresowań autora, opracowane na podstawie najnow szego piśmiennictwa, wystarczającego dla zilustrowania problemu. Ponadto kwartalnik publikuje krótkie noty, do 5 stron maszynopisu, informujące o nowych, interesujących osiągnięciach biochemii i nauk pokrewnych, oraz noty przybliżające historię badań w zakresie różnych dziedzin biochemii. Przekazanie artykułu do Redakcji jest równoznaczne z oświadczeniem, że nadesłana praca nie była i nie będzie publikowana w innym czasopiśmie, jeżeli zostanie ogłoszona w „Postępach Biochemii”. Autorzy artykułu odpowiadają za prawidłowość i ścisłość podanych informacji. Autorów obowiązuje korekta autorska. Koszty zmian tekstu w korekcie (poza poprawieniem błędów drukarskich) ponoszą autorzy. Artykuły honoruje się według obowiązujących stawek. Autorzy otrzymują bezpłatnie 25 odbitek swego artykułu; zamówienia na dodatkowe odbitki (płatne) należy zgłosić pisemnie odsyłając pracę po korekcie autorskiej. Redakcja prosi autorów o przestrzeganie następujących wskazówek: Forma maszynopisu: maszynopis pracy i wszelkie załączniki należy nadsyłać w dwu egzem plarzach. Maszynopis powinien być napisany jednostronnie, z podwójną interlinią, z marginesem ok. 4 cm po lewej i ok. 1 cm po prawej stronie; nie może zawierać więcej niż 60 znaków w jednym wierszu nie więcej niż 30 wierszy na stronie zgodnie z Normą Polską. -

(12) Patent Application Publication (10) Pub. No.: US 2006/0277.632 A1 Carr Et Al
US 20060277.632A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2006/0277.632 A1 Carr et al. (43) Pub. Date: Dec. 7, 2006 (54) METHODS FOR PRODUCTION OF CHITIN Publication Classification AND CHITOSAN (51) Int. Cl. AOIH I/00 (2006.01) (75) Inventors: Brian Carr, Raleigh, NC (US); Philip COSB 37/08 (2006.01) E. Hammer, Cary, NC (US) C7H 2L/04 (2006.01) CI2P 19/28 (2006.01) CI2N I/2 (2006.01) Correspondence Address: CI2N L/6 (2006.01) ALSTON & BIRD LLP CI2N 15/74 (2006.01) BANK OF AMERICA PLAZA (52) U.S. Cl. ...................... 800/284; 435/85; 435/252.33; 101 SOUTH TRYON STREET, SUITE 4000 435/254.3: 435/.484; 435/488; CHARLOTTE, NC 28280-4000 (US) 536/20, 536/23.2 (57) ABSTRACT Compositions and methods for producing chitin and chito Assignee: Athenix Corporation, Durham, NC (US) san are provided. The compositions comprise genetically (73) modified organisms, including fungi, yeast, bacterial and plant organisms that have been engineered to express het erologous genes involved in chitin and chitosan synthesis. (21) Appl. No.: 11/434,526 Microorganisms and plants that have been modified for production of chitin and/or chitosan within the vacuole of a cell are encompassed. Methods for production of chitin also (22) Filed: May 15, 2006 comprise culturing the genetically engineered organisms in conditions that allow for chitin production. Further methods include converting the chitin to chitosan by a chemical Related U.S. Application Data process. Production of chitosan also comprises culturing organisms that are genetically modified to produce chitosan (60) Provisional application No. -
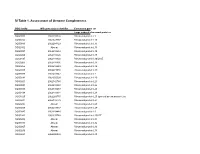
SI Table 1. Assessment of Genome Completeness
SI Table 1. Assessment of Genome Completeness COG family IMG gene object identifier Conserved gene set Large subunit ribosomal proteins COG0081 2062288324 Ribosomal protein L1 COG0244 2062347387 Ribosomal protein L10 COG0080 2062288323 Ribosomal protein L11 COG0102 Absent Ribosomal protein L13 COG0093 2062418832 Ribosomal protein L14 COG0200 2062418826 Ribosomal protein L15 COG0197 2062418838 Ribosomal protein L16/L10E COG0203 2062418836 Ribosomal protein L17 COG0256 2062418829 Ribosomal protein L18 COG0335 2062273558 Ribosomal protein L19 COG0090 2062418842 Ribosomal protein L2 COG0292 2062350539 Ribosomal protein L20 COG0261 2062142780 Ribosomal protein L21 COG0091 2062418840 Ribosomal protein L22 COG0089 2062138283 Ribosomal protein L23 COG0198 2062418834 Ribosomal protein L24 COG1825 2062269715 Ribosomal protein L25 (general stress protein Ctc) COG0211 2062142779 Ribosomal protein L27 COG0227 Absent Ribosomal protein L28 COG0255 2062418837 Ribosomal protein L29 COG0087 2062154483 Ribosomal protein L3 COG1841 2062335748 Ribosomal protein L30/L7E COG0254 Absent Ribosomal protein L31 COG0333 Absent Ribosomal protein L32 COG0267 Absent Ribosomal protein L33 COG0230 Absent Ribosomal protein L34 COG0291 2062350538 Ribosomal protein L35 COG0257 Absent Ribosomal protein L36 COG0088 2062138282 Ribosomal protein L4 COG0094 2062418833 Ribosomal protein L5 COG0097 2062418830 Ribosomal protein L6P/L9E COG0222 2062288326 Ribosomal protein L7/L12 COG0359 2062209880 Ribosomal protein L9 Small subunit ribosomal proteins COG0539 Absent Ribosomal protein