Lignin Intermediates Lead to Phenyl Acid Formation and Microbial
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Archaea, Bacteria and Termite, Nitrogen Fixation and Sustainable Plants Production
Sun W et al . (2021) Notulae Botanicae Horti Agrobotanici Cluj-Napoca Volume 49, Issue 2, Article number 12172 Notulae Botanicae Horti AcademicPres DOI:10.15835/nbha49212172 Agrobotanici Cluj-Napoca Re view Article Archaea, bacteria and termite, nitrogen fixation and sustainable plants production Wenli SUN 1a , Mohamad H. SHAHRAJABIAN 1a , Qi CHENG 1,2 * 1Chinese Academy of Agricultural Sciences, Biotechnology Research Institute, Beijing 100081, China; [email protected] ; [email protected] 2Hebei Agricultural University, College of Life Sciences, Baoding, Hebei, 071000, China; Global Alliance of HeBAU-CLS&HeQiS for BioAl-Manufacturing, Baoding, Hebei 071000, China; [email protected] (*corresponding author) a,b These authors contributed equally to the work Abstract Certain bacteria and archaea are responsible for biological nitrogen fixation. Metabolic pathways usually are common between archaea and bacteria. Diazotrophs are categorized into two main groups namely: root- nodule bacteria and plant growth-promoting rhizobacteria. Diazotrophs include free living bacteria, such as Azospirillum , Cupriavidus , and some sulfate reducing bacteria, and symbiotic diazotrophs such Rhizobium and Frankia . Three types of nitrogenase are iron and molybdenum (Fe/Mo), iron and vanadium (Fe/V) or iron only (Fe). The Mo-nitrogenase have a higher specific activity which is expressed better when Molybdenum is available. The best hosts for Rhizobium legumiosarum are Pisum , Vicia , Lathyrus and Lens ; Trifolium for Rhizobium trifolii ; Phaseolus vulgaris , Prunus angustifolia for Rhizobium phaseoli ; Medicago, Melilotus and Trigonella for Rhizobium meliloti ; Lupinus and Ornithopus for Lupini, and Glycine max for Rhizobium japonicum . Termites have significant key role in soil ecology, transporting and mixing soil. Termite gut microbes supply the enzymes required to degrade plant polymers, synthesize amino acids, recycle nitrogenous waste and fix atmospheric nitrogen. -

Desulfuribacillus Alkaliarsenatis Gen. Nov. Sp. Nov., a Deep-Lineage
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central Extremophiles (2012) 16:597–605 DOI 10.1007/s00792-012-0459-7 ORIGINAL PAPER Desulfuribacillus alkaliarsenatis gen. nov. sp. nov., a deep-lineage, obligately anaerobic, dissimilatory sulfur and arsenate-reducing, haloalkaliphilic representative of the order Bacillales from soda lakes D. Y. Sorokin • T. P. Tourova • M. V. Sukhacheva • G. Muyzer Received: 10 February 2012 / Accepted: 3 May 2012 / Published online: 24 May 2012 Ó The Author(s) 2012. This article is published with open access at Springerlink.com Abstract An anaerobic enrichment culture inoculated possible within a pH range from 9 to 10.5 (optimum at pH with a sample of sediments from soda lakes of the Kulunda 10) and a salt concentration at pH 10 from 0.2 to 2 M total Steppe with elemental sulfur as electron acceptor and for- Na? (optimum at 0.6 M). According to the phylogenetic mate as electron donor at pH 10 and moderate salinity analysis, strain AHT28 represents a deep independent inoculated with sediments from soda lakes in Kulunda lineage within the order Bacillales with a maximum of Steppe (Altai, Russia) resulted in the domination of a 90 % 16S rRNA gene similarity to its closest cultured Gram-positive, spore-forming bacterium strain AHT28. representatives. On the basis of its distinct phenotype and The isolate is an obligate anaerobe capable of respiratory phylogeny, the novel haloalkaliphilic anaerobe is suggested growth using elemental sulfur, thiosulfate (incomplete as a new genus and species, Desulfuribacillus alkaliar- T T reduction) and arsenate as electron acceptor with H2, for- senatis (type strain AHT28 = DSM24608 = UNIQEM mate, pyruvate and lactate as electron donor. -

Antonie Van Leeuwenhoek Journal of Microbiology
Antonie van Leeuwenhoek Journal of Microbiology Kroppenstedtia pulmonis sp. nov. and Kroppenstedtia sanguinis sp. nov., isolated from human patients --Manuscript Draft-- Manuscript Number: ANTO-D-15-00548R1 Full Title: Kroppenstedtia pulmonis sp. nov. and Kroppenstedtia sanguinis sp. nov., isolated from human patients Article Type: Original Article Keywords: Kroppenstedtia species, Kroppenstedtia pulmonis, Kroppenstedtia sanguinis, polyphasic taxonomy, 16S rRNA gene, thermoactinomycetes Corresponding Author: Melissa E Bell, MS Centers for Disease Control and Prevention Atlanta, Georgia UNITED STATES Corresponding Author Secondary Information: Corresponding Author's Institution: Centers for Disease Control and Prevention Corresponding Author's Secondary Institution: First Author: Melissa E Bell, MS First Author Secondary Information: Order of Authors: Melissa E Bell, MS Brent A. Lasker, PhD Hans-Peter Klenk, PhD Lesley Hoyles, PhD Catherine Spröer Peter Schumann June Brown Order of Authors Secondary Information: Funding Information: Abstract: Three human clinical strains (W9323T, X0209T and X0394) isolated from lung biopsy, blood and cerebral spinal fluid, respectively, were characterized using a polyphasic taxonomic approach. Comparative analysis of the 16S rRNA gene sequences showed the three strains belonged to two novel branches within the genus Kroppenstedtia: 16S rRNA gene sequence analysis of W9323T showed closest sequence similarity to Kroppenstedtia eburnea JFMB-ATE T (95.3 %), Kroppenstedtia guangzhouensis GD02T (94.7 %) and strain X0209T (94.6 %); sequence analysis of strain X0209T showed closest sequence similarity to K. eburnea JFMB-ATE T (96.4 %) and K. guangzhouensis GD02T (96.0 %). Strains X0209T and X0394 were 99.9 % similar to each other by 16S rRNA gene sequence analysis. The DNA-DNA relatedness was 94.6 %, confirming that X0209T and X0394 belong to the same species. -

Revised Supplement 1: Reference List for Figure 1
Revised Supplement 1: Reference list for Figure 1. Manuscript title: Process disturbances in agricultural biogas production – causes, mechanisms and effects on the biogas microbiome: A review Susanne Theuerl 1,*, Johanna Klang 1, Annette Prochnow 1,2 1 Leibniz Institute for Agricultural Engineering and Bioeconomy, Max-Exth-Allee 100, 14469 Potsdam, Germany, [email protected] (ST), [email protected] (JK), [email protected] (AP) 2 Humboldt-Universität zu Berlin, Albrecht-Daniel-Thaer-Institute for Agricultural and Horticultural Sciences, Hinter der Reinhardtstr. 6-8, 10115 Berlin, Germany * Correspondence: [email protected] Tel.: +49-331-5699-900 References of Figure 1 1Abt et al. 2010, 2Parizzi et al. 2012, 3Hahnke et al. 2016 and Tomazetto et al. 2018, 4Ueki et al. 2006 and Gronow et al. 2011, 5Grabowski et al. 2005, 6Chen and Dong 2005, 7Avgustin et al. 1997 and Purushe et al. 2010, 8Yamada et al. 2006 and Matsuura et al. 2015, 9Yamada et al. 2007 and Matsuura et al. 2015, 10Sun et al. 2016, 11Suen et al. 2011, 12Hahnke et al. 2014 and Tomazetto et al. 2016, 13Mechichi et al. 1999, 14Koeck et al. 2015a and 2015b, 15Tomazetto et al. 2017, 16Fonknechten et al. 2010, 17Chen et al. 2010, 18Nishiyama et al. 2009, 19Sieber et al. 2010, 20Plerce et al. 2008, 21Westerholm et al. 2011 and Müller et al. 2015, 22Ueki et al. 2014, 23Jackson et al. 1999 and McInerney et al. 2007, 24Ma et al. 2017, 25Harmsen et al. 1998 and Plugge et al. 2012, 26Menes and Muxi 2002, Mavromatis et al. 2013 and Hania et al. -

Lascolabacillus Massiliensis'': a New Species Isolated
NEW MICROBES IN HUMANS “Lascolabacillus massiliensis”: a new consent, and the agreement of the National Ethics Committee of Senegal and the local ethics committee of the IFR48 species isolated from the human gut (Marseille, France) were obtained under numbers 11-017 and 09-022, respectively. The initial growth was obtained after 10 days of culture in a 5% sheep blood-enriched sheep rumen M. Beye, S. Bakour, S. I. Traore, D. Raoult and P.-E. Fournier medium in aerobic atmosphere at 37°C. The bacterium was Unité de Recherche en Maladies Infectieuses et Tropicales Emergentes, sub-cultured on 5% sheep blood-enriched Columbia agar Institut Hospitalo-Universitaire Méditerranée-Infection, Aix-Marseille (bioMérieux, Marcy l’Etoile, France) and grew in 24 hours at Université, Faculté de Médecine, Marseille cedex 5, France 37°C in both aerobic and anaerobic conditions. Agar-grown colonies were pale grey and 1.5 mm in diameter. Bacterial cells were Gram-negative, rod-shaped and polymorphic, ranginginlengthfrom1.5to10μm. Strain SIT8 was catalase- Abstract and oxidase-negative. The 16S rRNA gene was sequenced using the fD1-rP2 primers as previously described, using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, We report here the main characteristics of “Lascolabacillus France). Strain SIT8 exhibited a 94.14% sequence identity with massiliensis” strain SIT8 (CSUR P1560) that was isolated from the Proteiniphilum acetatigenes strain TB107 (GenBank accession stool of a healthy 28-month-old boy. NR_043154), the phylogenetically closest species with New Microbes and New Infections © 2016 The Authors. Published standing in nomenclature (Fig. 1), which putatively classifies it by Elsevier Ltd on behalf of European Society of Clinical as a member of a new genus within the family Porphyr- Microbiology and Infectious Diseases. -
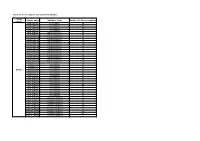
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

Impact of Cropping Systems, Soil Inoculum, and Plant Species Identity on Soil Bacterial Community Structure
Impact of Cropping Systems, Soil Inoculum, and Plant Species Identity on Soil Bacterial Community Structure Authors: Suzanne L. Ishaq, Stephen P. Johnson, Zach J. Miller, Erik A. Lehnhoff, Sarah Olivo, Carl J. Yeoman, and Fabian D. Menalled The final publication is available at Springer via http://dx.doi.org/10.1007/s00248-016-0861-2. Ishaq, Suzanne L. , Stephen P. Johnson, Zach J. Miller, Erik A. Lehnhoff, Sarah Olivo, Carl J. Yeoman, and Fabian D. Menalled. "Impact of Cropping Systems, Soil Inoculum, and Plant Species Identity on Soil Bacterial Community Structure." Microbial Ecology 73, no. 2 (February 2017): 417-434. DOI: 10.1007/s00248-016-0861-2. Made available through Montana State University’s ScholarWorks scholarworks.montana.edu Impact of Cropping Systems, Soil Inoculum, and Plant Species Identity on Soil Bacterial Community Structure 1,2 & 2 & 3 & 4 & Suzanne L. Ishaq Stephen P. Johnson Zach J. Miller Erik A. Lehnhoff 1 1 2 Sarah Olivo & Carl J. Yeoman & Fabian D. Menalled 1 Department of Animal and Range Sciences, Montana State University, P.O. Box 172900, Bozeman, MT 59717, USA 2 Department of Land Resources and Environmental Sciences, Montana State University, P.O. Box 173120, Bozeman, MT 59717, USA 3 Western Agriculture Research Center, Montana State University, Bozeman, MT, USA 4 Department of Entomology, Plant Pathology and Weed Science, New Mexico State University, Las Cruces, NM, USA Abstract Farming practices affect the soil microbial commu- then individual farm. Living inoculum-treated soil had greater nity, which in turn impacts crop growth and crop-weed inter- species richness and was more diverse than sterile inoculum- actions. -

Melghirimyces Thermohalophilus Sp. Nov., a Thermoactinomycete Isolated from an Algerian Salt Lake
International Journal of Systematic and Evolutionary Microbiology (2013), 63, 1717–1722 DOI 10.1099/ijs.0.043760-0 Melghirimyces thermohalophilus sp. nov., a thermoactinomycete isolated from an Algerian salt lake Ammara Nariman Addou,1,2 Peter Schumann,3 Cathrin Spro¨er,3 Amel Bouanane-Darenfed,2 Samia Amarouche-Yala,4 Hocine Hacene,2 Jean-Luc Cayol1 and Marie-Laure Fardeau1 Correspondence 1Aix-Marseille Universite´, Universite´ du Sud Toulon-Var, CNRS/INSU, IRD, MIO, UM 110, 13288 Marie-Laure Fardeau Marseille Cedex 09, France [email protected] 2Laboratoire de Biologie Cellulaire et Mole´culaire (e´quipe de Microbiologie), Universite´ des sciences et de la technologie, Houari Boume´die`nne, Bab Ezzouar, Algiers, Algeria 3Leibniz Institut DSMZ – Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Inhoffenstraße 7B, 38124 Braunschweig, Germany 4Centre de Recherche Nucle´aire d’Alger (CRNA), Algeria A novel filamentous bacterium, designated Nari11AT, was isolated from soil collected from a salt lake named Chott Melghir, located in north-eastern Algeria. The strain is an aerobic, halophilic, thermotolerant, Gram-stain-positive bacterium, growing at NaCl concentrations between 5 and 20 % (w/v) and at 43–60 6C and pH 5.0–10.0. The major fatty acids were iso-C15 : 0, anteiso- C15 : 0 and iso-C17 : 0. The DNA G+C content was 53.4 mol%. LL-Diaminopimelic acid was the diamino acid of the peptidoglycan. The major menaquinone was MK-7, but MK-6 and MK-8 were also present in trace amounts. The polar lipid profile consisted of phosphatidylglycerol, diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylmonomethylethanolamine and three unidentified phospholipids. -

Characterization of Cucumber Fermentation Spoilage Bacteria by Enrichment Culture and 16S Rdna Cloning
Characterization of Cucumber Fermentation Spoilage Bacteria by Enrichment Culture and 16S rDNA Cloning Fred Breidt, Eduardo Medina, Doria Wafa, Ilenys P´erez-D´ıaz, Wendy Franco, Hsin-Yu Huang, Suzanne D. Johanningsmeier, and Jae Ho Kim Abstract: Commercial cucumber fermentations are typically carried out in 40000 L fermentation tanks. A secondary fermentation can occur after sugars are consumed that results in the formation of acetic, propionic, and butyric acids, concomitantly with the loss of lactic acid and an increase in pH. Spoilage fermentations can result in significant economic loss for industrial producers. The microbiota that result in spoilage remain incompletely defined. Previous studies have implicated yeasts, lactic acid bacteria, enterobacteriaceae, and Clostridia as having a role in spoilage fermentations. We report that Propionibacterium and Pectinatus isolates from cucumber fermentation spoilage converted lactic acid to propionic acid, increasing pH. The analysis of 16S rDNA cloning libraries confirmed and expanded the knowledge gained from previous studies using classical microbiological methods. Our data show that Gram-negative anaerobic bacteria supersede Gram-positive Fermincutes species after the pH rises from around 3.2 to pH 5, and propionic and butyric acids are produced. Characterization of the spoilage microbiota is an important first step in efforts to prevent cucumber fermentation spoilage. Keywords: pickled vegetables, Pectinatus, Propionibacteria, secondary cucumber fermentation, spoilage M: Food Microbiology Practical Application: An understanding of the microorganisms that cause commercial cucumber fermentation spoilage & Safety may aid in developing methods to prevent the spoilage from occurring. Introduction cucumbers fermented at 2.3% NaCl (Fleming and others 1989). Commercial cucumber fermentations are typically carried out In this fermentation tank, the initial lactic acid fermentation was in large 40000 L outdoor tanks (reviewed by Breidt and others completed within 2 wk, with 1.2% lactic acid formed (pH 3.6) 2007). -

Microbial Communities Associated with Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully USDA-ARS, [email protected]
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications: Department of Entomology Entomology, Department of 2017 Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully USDA-ARS, [email protected] Kristina Friesen USDA-ARS, [email protected] Brian Wienhold USDA-ARS, [email protected] Lisa M. Durso USDA-ARS, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/entomologyfacpub Part of the Entomology Commons Scully, Erin; Friesen, Kristina; Wienhold, Brian; and Durso, Lisa M., "Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their eD velopmental Substrates" (2017). Faculty Publications: Department of Entomology. 502. http://digitalcommons.unl.edu/entomologyfacpub/502 This Article is brought to you for free and open access by the Entomology, Department of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Faculty Publications: Department of Entomology by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Annals of the Entomological Society of America, 110(1), 2017, 61–72 doi: 10.1093/aesa/saw087 Special Collection: Filth Fly–Microbe Interactions Research article Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully,1 Kristina Friesen,2,3 Brian Wienhold,2 and Lisa M. Durso2 1USDA, ARS, Stored Product -

Page 1 of 41 RSC Advances
RSC Advances This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. This Accepted Manuscript will be replaced by the edited, formatted and paginated article as soon as this is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/advances Page 1 of 41 RSC Advances 1 Regulation of acidogenic metabolism towards enhanced short chain fatty acids biosynthesis 2 from waste: Metagenomic Profiling 3 4 Omprakash Sarkar, A. Naresh Kumar, Shikha Dahiya, K.Vamshi Krishna, Dileep Kumar 5 Yeruva, S.Venkata Mohan* 6 Bioengineering and Environmental Sciences (BEES), CSIR-Indian Institute of Chemical 7 Technology (CSIR-IICT), Hyderabad 500 007, India 8 *E-mail: [email protected]; Tel: 0091-40-27161765 9 10 Abstract 11 Short chain carboxylic (volatile fatty) acids (VFA) production in mixed microbiomes is majorly 12 limited by the prevalence of the methanogenic bacteria and availability of substrate from waste 13 to the biocatalyst during the fermentation process. -

The Genus Geobacillus
The Genus Geobacillus Introduction and Strain Catalog polymyxa macerans popilliae brevis Paenibacillus pantothenticus agri Virgibacillus laterosporus Brevibacillus cycloheptanicus Alicyclobacillus salexigens acidocaldarius Salibacillus marismortui halotolerans acidoterrestris Gracilibacillus thermoaerophilus Aneurinibacillus subtilis migulanus Bacillus cereus Geobacillus Ureibacillus kaustophilus badius stearothermophilus subterraneus halophilus thermosphaericus terreneus Bacillus Genetic Stock Center Catalog of Strains 7th Edition, Volume 3 Bacillus Genetic Stock Center Catalog of Strains, Seventh Edition Volume 3: The Genus Geobacillus © 2001 Permission is given to copy this material or portions of it freely, provided that no alterations are made and that this copyright page is included. Daniel R. Zeigler, Ph.D. The Bacillus Genetic Stock Center Department of Biochemistry The Ohio State University 484 West Twelfth Avenue Columbus, Ohio 43210 Disclaimer: The information in this catalog is believed to be correct. Due to the dynamic nature of the scientific process and to normal human limitations in dealing with such a large amount of data, however, some undetected errors may persist. Users bear the responsibility of verifying any important data before making a significant investment of time or other physical or financial resources. Cover: Phylogenetic tree of the genus Bacillus and closely related genera, including the new genus Geobacillus (Nazina, 2001). 16S rRNA gene sequences were obtained from GenBank for the species represented on the tree. After they were manually trimmed to the same length, they were aligned with ClustalW to create a Phylip distance matrix. The Neighbor program from the Phylip suite was used to generate a UPGMA tree, which was visualized with TreeView32. I used MacroMedia Freehand and Microsoft Image Composer to edit the tree image.