Gene Section Review
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FIP1L1 Gene FIP1 Like 1 (S
FIP1L1 gene FIP1 like 1 (S. cerevisiae) Normal Function The FIP1L1 gene provides instructions for making part of a protein complex named cleavage and polyadenylation specificity factor (CPSF). This complex of proteins plays an important role in processing molecules called messenger RNAs (mRNAs), which serve as the genetic blueprints for making proteins. The CPSF protein complex helps add a string of the RNA building block adenine to the mRNA, creating a polyadenine tail or poly(A) tail. The poly(A) tail is important for stability of the mRNA and for protein production from the blueprint. Health Conditions Related to Genetic Changes PDGFRA-associated chronic eosinophilic leukemia A deletion of genetic material from chromosome 4 brings together part of the FIP1L1 gene and part of another gene called PDGFRA, creating the FIP1L1-PDGFRA fusion gene. This mutation is a somatic mutation, which means it is acquired during a person's lifetime and is present only in certain cells. This fusion gene causes PDGFRA- associated chronic eosinophilic leukemia, which is a type of blood cell cancer characterized by an increased number of eosinophils, a type of white blood cell involved in allergic reactions. The FIP1L1-PDGFRA protein produced from the fusion gene has the function of the normal PDGFRA protein, which stimulates signaling pathways inside the cell that control many important cellular processes, such as cell growth and division (proliferation) and cell survival. Unlike the normal PDGFRA protein, however, the FIP1L1-PDGFRA protein is constantly turned on (constitutively activated), which means the cells are always receiving signals to proliferate. When the FIP1L1-PDGFRA fusion gene occurs in blood cell precursors, the growth of eosinophils (and occasionally other blood cells) is poorly controlled, leading to PDGFRA-associated chronic eosinophilic leukemia. -

R-Loop-Mediated Genome Instability in Mrna Cleavage and Polyadenylation Mutants
Downloaded from genesdev.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants Peter C. Stirling,1,3 Yujia A. Chan,1,3 Sean W. Minaker,1,3 Maria J. Aristizabal,2 Irene Barrett,1 Payal Sipahimalani,1 Michael S. Kobor,2 and Philip Hieter1,4 1Michael Smith Laboratories, University of British Columbia, Vancouver, British Columbia V6T1Z4, Canada; 2Centre for Molecular Medicine and Therapeutics, Vancouver, British Columbia V5Z4H4, Canada Genome instability via RNA:DNA hybrid-mediated R loops has been observed in mutants involved in various aspects of transcription and RNA processing. The prevalence of this mechanism among essential chromosome instability (CIN) genes remains unclear. In a secondary screen for increased Rad52 foci in CIN mutants, representing ~25% of essential genes, we identified seven essential subunits of the mRNA cleavage and polyadenylation (mCP) machinery. Genome-wide analysis of fragile sites by chromatin immunoprecipitation (ChIP) and microarray (ChIP–chip) of phosphorylated H2A in these mutants supported a transcription-dependent mechanism of DNA damage characteristic of R loops. In parallel, we directly detected increased RNA:DNA hybrid formation in mCP mutants and demonstrated that CIN is suppressed by expression of the R-loop-degrading enzyme RNaseH. To investigate the conservation of CIN in mCP mutants, we focused on FIP1L1, the human ortholog of yeast FIP1, a conserved mCP component that is part of an oncogenic fusion in eosinophilic leukemia. We found that truncation fusions of yeast FIP1 analogous to those in cancer cause loss of function and that siRNA knockdown of FIP1L1 in human cells increases DNA damage and chromosome breakage. -

Hypereosinophilia in Granular Acute Bcell Lymphoblastic Leukemia With
bs_bs_banner Microscopic polyangiitis 543 and had been diagnosed with normocytic anemia.At that time, she References received treatment with an iron preparation because of a low 1 Ozen S, Ruperto N, Dillon MJ et al. EULAR/PReS endorsed con- serum iron level despite her normal ferritin level. We estimate that sensus criteria for the classification of childhood vasculitides. Ann. the patient already had mild alveolar hemorrhage at that time and Rheum. Dis. 2006; 65: 936–41. the bleeding later healed spontaneously. Among the cases of this 2 Vanoni F, Bettinelli A, Keller F, Bianchetti MG, Simonetti GD. disease reported from Japan, the disease tended to develop more Vasculitides associated with IgG antineutrophil cytoplasmic autoan- tibodies in childhood. Pediatr. Nephrol. 2010; 25: 205–12. frequently in the spring. In our patient, the disease developed twice 3 Kobayashi S, Inokuma S. Intrapulmonary hemorrhage in collagen- (both times in June), suggesting the possibility that infection vascular disease includes a spectrum of underlying conditions. triggered the disease onset. Although alveolar hemorrhage sub- Intern. Med. 2009; 48: 891–7. sided spontaneously in this case, lung fibrosis may develop in the 4 Hattori M, Kurayama H, Koitabashi Y. Antineutrophil cytoplasmic future if alveolar bleeding develops repeatedly and its diagnosis is autoantibody-associated glomerulonephritis in children. J. Am. Soc. Nephrol. 2001; 12: 1493–500. delayed. In the present case, treatment was started immediately 5 Peco-Antic A, Bonaci-Nikolic B, Basta-Jovanovic G et al. Child- after diagnosis and rapid aggravation of her renal function was hood microscopic polyangiitis associated with MPO-ANCA. prevented. When dealing with cases in whom unexplained nor- Pediatr. -

PRODUCT SPECIFICATION Prest Antigen FIP1L1 Product Datasheet
PrEST Antigen FIP1L1 Product Datasheet PrEST Antigen PRODUCT SPECIFICATION Product Name PrEST Antigen FIP1L1 Product Number APrEST79770 Gene Description factor interacting with PAPOLA and CPSF1 Alternative Gene DKFZp586K0717 Names Corresponding Anti-FIP1L1 (HPA037475) Antibodies Description Recombinant protein fragment of Human FIP1L1 Amino Acid Sequence Recombinant Protein Epitope Signature Tag (PrEST) antigen sequence: KANSSVGKWQDRYGRAESPDLRRLPGAIDVIGQTITISRVEGRRRANENS NIQVLSERSATEVDNNFSKPPPFFPPGAPPT Fusion Tag N-terminal His6ABP (ABP = Albumin Binding Protein derived from Streptococcal Protein G) Expression Host E. coli Purification IMAC purification Predicted MW 27 kDa including tags Usage Suitable as control in WB and preadsorption assays using indicated corresponding antibodies. Purity >80% by SDS-PAGE and Coomassie blue staining Buffer PBS and 1M Urea, pH 7.4. Unit Size 100 µl Concentration Lot dependent Storage Upon delivery store at -20°C. Avoid repeated freeze/thaw cycles. Notes Gently mix before use. Optimal concentrations and conditions for each application should be determined by the user. Product of Sweden. For research use only. Not intended for pharmaceutical development, diagnostic, therapeutic or any in vivo use. No products from Atlas Antibodies may be resold, modified for resale or used to manufacture commercial products without prior written approval from Atlas Antibodies AB. Warranty: The products supplied by Atlas Antibodies are warranted to meet stated product specifications and to conform to label descriptions when used and stored properly. Unless otherwise stated, this warranty is limited to one year from date of sales for products used, handled and stored according to Atlas Antibodies AB's instructions. Atlas Antibodies AB's sole liability is limited to replacement of the product or refund of the purchase price. -
Letters to the Editor
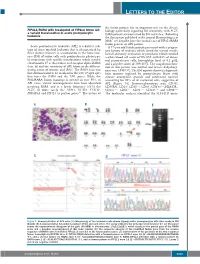
LETTERS TO THE EDITOR the fusion partner has an important role on the disease FIP1L1/RARA with breakpoint at FIP1L1 intron 13: biology particularly regarding RA sensitivity, with PLZF- a variant translocation in acute promyelocytic 4 leukemia RARA patients characterized by RA resistance. Following the description published in the journal Haematologica in 2008, 5 we describe here the second case of FIP1L1/RARA fusion gene in an APL patient. Acute promyelocytic leukemia (APL) is a distinct sub - A 77-year old female patient presented with a progres - type of acute myeloid leukemia that is characterized by sive history of asthenia which lasted for several weeks. three distinct features: i) accumulation in the bone mar - Initial laboratory evaluation of peripheral blood revealed row (BM) of tumor cells with promyelocytic phenotype; a white blood cell count of 59 ¥10 9/L with 84% of abnor - ii) association with specific translocations which involve mal promyelocyte cells, hemoglobin level of 9.2 g/dL, chromosome 17 at the retinoic acid receptor alpha ( RARA ) and a platelet count of 109 ¥10 9/L. The coagulation func - locus ; iii) and the sensitivity of APL blasts to the differen - tion in this patient was normal and lactate dehydroge - 1 tiating action of retinoic acid (RA). The RARA locus was nase was 1,938 U/L. The BM aspirate showed a hypercel - first demonstrated to be involved in the t(15;17)(q22;q21) lular marrow replaced by promyelocyte blasts with that fuses the RARA and the PML genes. While the intense azurophilic granule and prominent nucleoli PML/RARA fusion transcript is present in over 95% of accounting for 93% of all nucleated cells, suggestive of APL cases, variant rearrangements have been identified APL (Figure 1A). -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

CPTC-CDK1-1 (CAB079974) Immunohistochemistry
CPTC-CDK1-1 (CAB079974) Uniprot ID: P06493 Protein name: CDK1_HUMAN Full name: Cyclin-dependent kinase 1 Tissue specificity: Isoform 2 is found in breast cancer tissues. Function: Plays a key role in the control of the eukaryotic cell cycle by modulating the centrosome cycle as well as mitotic onset; promotes G2-M transition, and regulates G1 progress and G1-S transition via association with multiple interphase cyclins. Required in higher cells for entry into S-phase and mitosis. Phosphorylates PARVA/actopaxin, APC, AMPH, APC, BARD1, Bcl-xL/BCL2L1, BRCA2, CALD1, CASP8, CDC7, CDC20, CDC25A, CDC25C, CC2D1A, CENPA, CSNK2 proteins/CKII, FZR1/CDH1, CDK7, CEBPB, CHAMP1, DMD/dystrophin, EEF1 proteins/EF-1, EZH2, KIF11/EG5, EGFR, FANCG, FOS, GFAP, GOLGA2/GM130, GRASP1, UBE2A/hHR6A, HIST1H1 proteins/histone H1, HMGA1, HIVEP3/KRC, LMNA, LMNB, LMNC, LBR, LATS1, MAP1B, MAP4, MARCKS, MCM2, MCM4, MKLP1, MYB, NEFH, NFIC, NPC/nuclear pore complex, PITPNM1/NIR2, NPM1, NCL, NUCKS1, NPM1/numatrin, ORC1, PRKAR2A, EEF1E1/p18, EIF3F/p47, p53/TP53, NONO/p54NRB, PAPOLA, PLEC/plectin, RB1, TPPP, UL40/R2, RAB4A, RAP1GAP, RCC1, RPS6KB1/S6K1, KHDRBS1/SAM68, ESPL1, SKI, BIRC5/survivin, STIP1, TEX14, beta-tubulins, MAPT/TAU, NEDD1, VIM/vimentin, TK1, FOXO1, RUNX1/AML1, SAMHD1, SIRT2 and RUNX2. CDK1/CDC2-cyclin-B controls pronuclear union in interphase fertilized eggs. Essential for early stages of embryonic development. During G2 and early mitosis, CDC25A/B/C-mediated dephosphorylation activates CDK1/cyclin complexes which phosphorylate several substrates that trigger at least centrosome separation, Golgi dynamics, nuclear envelope breakdown and chromosome condensation. Once chromosomes are condensed and aligned at the metaphase plate, CDK1 activity is switched off by WEE1- and PKMYT1-mediated phosphorylation to allow sister chromatid separation, chromosome decondensation, reformation of the nuclear envelope and cytokinesis. -

Downloaded the “Top Edge” Version
bioRxiv preprint doi: https://doi.org/10.1101/855338; this version posted December 6, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Drosophila models of pathogenic copy-number variant genes show global and 2 non-neuronal defects during development 3 Short title: Non-neuronal defects of fly homologs of CNV genes 4 Tanzeen Yusuff1,4, Matthew Jensen1,4, Sneha Yennawar1,4, Lucilla Pizzo1, Siddharth 5 Karthikeyan1, Dagny J. Gould1, Avik Sarker1, Yurika Matsui1,2, Janani Iyer1, Zhi-Chun Lai1,2, 6 and Santhosh Girirajan1,3* 7 8 1. Department of Biochemistry and Molecular Biology, Pennsylvania State University, 9 University Park, PA 16802 10 2. Department of Biology, Pennsylvania State University, University Park, PA 16802 11 3. Department of Anthropology, Pennsylvania State University, University Park, PA 16802 12 4 contributed equally to work 13 14 *Correspondence: 15 Santhosh Girirajan, MBBS, PhD 16 205A Life Sciences Building 17 Pennsylvania State University 18 University Park, PA 16802 19 E-mail: [email protected] 20 Phone: 814-865-0674 21 1 bioRxiv preprint doi: https://doi.org/10.1101/855338; this version posted December 6, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 22 ABSTRACT 23 While rare pathogenic copy-number variants (CNVs) are associated with both neuronal and non- 24 neuronal phenotypes, functional studies evaluating these regions have focused on the molecular 25 basis of neuronal defects. -

ORIGINAL ARTICLE the Severity of FIP1L1–PDGFRA-Positive Chronic
Leukemia (2007) 21, 2428–2432 & 2007 Nature Publishing Group All rights reserved 0887-6924/07 $30.00 www.nature.com/leu ORIGINAL ARTICLE The severity of FIP1L1–PDGFRA-positive chronic eosinophilic leukaemia is associated with polymorphic variation at the IL5RA locus S Burgstaller1, S Kreil1, K Waghorn1, G Metzgeroth2, C Preudhomme3, K Zoi4, H White1, D Cilloni5, C Zoi4, F Brito-Babapulle6, C Walz2, A Reiter2 and NCP Cross1 1Wessex Regional Genetics Laboratory, University of Southampton, Salisbury, UK; 2III Medizinische Universita¨tsklinik, Medizinische Fakulta¨t der Universita¨t Mannheim, Heidelberg, Germany; 3Laboratoire d’Hematologie A, CHU Lille, Lille, France; 4Foundation of Biomedical Research, Academy of Athens, Athens, Greece; 5Department of Clinical and Biological Sciences, University of Turin, Turin, Italy and 6Department of Haematology, Ealing Hospital, London, UK We have investigated the hypothesis that constitutional genetic Understanding of the myeloproliferative subtype of HES was variation in IL-5 signalling may be associated with the greatly advanced by the finding of the FIP1L1–PDGFRA fusion, development or severity of FIP1L1–PDGFRA-positive chronic formed by a cytogenetically cryptic 800 kb interstitial deletion at eosinophilic leukaemia (CEL) in humans. We genotyped six 2 single-nucleotide polymorphisms (SNP) within or close to the chromosome band 4q12. Although initially described in more IL5RA or IL5 genes in 82 patients with FIP1L1–PDGFRA- than 50% of cases with idiopathic HES, subsequent studies positive CEL plus, as controls, healthy individuals (n ¼ 100), estimated the prevalence to be lower at 3À17%.3–5 Detection of patients with FIP1L1–PDGFRA-negative eosinophilia (n ¼ 100) FIP1L1–PDGFRA by reverse transcription-PCR or fluorescent in or patients with chronic myeloid leukaemia (CML) (n ¼ 100). -

FIP1L1‑PDGFRA Fusion‑Negative Hypereosinophilic Syndrome with Uncommon Cardiac Involvement Responding to Imatinib Treatment: a Case Report
MOLECULAR AND CLINICAL ONCOLOGY 9: 35-39, 2018 FIP1L1‑PDGFRA fusion‑negative hypereosinophilic syndrome with uncommon cardiac involvement responding to imatinib treatment: A case report AMANDA SANTOS DAL BERTO1, RICARDO HOHMANN CAMIÑA1, EDUARDO SILVA MACHADO1-3 and ANTUANI RAFAEL BAPTISTELLA1,2,4,5 1Santa Terezinha University Hospital; 2University of West Santa Catarina; 3Department of Clinical Oncology, Santa Terezinha University Hospital; 4Oncology Research Group of Santa Terezinha University Hospital /University of West Santa Catarina; 5Post Graduation Program in Bioscience and Health/University of West Santa Catarina, Joaçaba, Santa Catarina 89600-000, Brazil Received April 17, 2018; Accepted May 21, 2018 DOI: 10.3892/mco.2018.1637 Abstract. Hypereosinophilic syndrome is a rare, chronic therapeutic possibilities were evaluated due to the significant hematological disease characterized by a persistently elevated progression of the disease, and it was decided to attempt the eosinophil count exceeding 1.5x109/l, following the exclusion use of imatinib, despite its use being preferably recommended of other potential etiologies. The systemic involvement of the for FIPIL1-PDGFRA-positive patients. The patient exhibited an disease causes tissue damage through eosinophil infiltration, evident and immediate response to imatinib, with normalization and may affect various organs; cardiac complications are of the eosinophil count within 24 h of the first dose, which was observed in 50-60% of cases, which are predominately attrib- maintained for at least the next 19 months. This clinical presen- uted to endomyocardial fibrosis. The treatment is based initially tation is uncommon, as patients negative for FIPIL1-PDGFRA on determining the presence of the FIP1L1-PDGFRA fusion. fusion do not frequently respond to imatinib treatment, and Patients with positive results for this mutation tend to achieve symptomatic heart failure usually appears in the third stage of a complete response with imatinib treatment, which is thus disease progression. -

Strand Breaks for P53 Exon 6 and 8 Among Different Time Course of Folate Depletion Or Repletion in the Rectosigmoid Mucosa
SUPPLEMENTAL FIGURE COLON p53 EXONIC STRAND BREAKS DURING FOLATE DEPLETION-REPLETION INTERVENTION Supplemental Figure Legend Strand breaks for p53 exon 6 and 8 among different time course of folate depletion or repletion in the rectosigmoid mucosa. The input of DNA was controlled by GAPDH. The data is shown as ΔCt after normalized to GAPDH. The higher ΔCt the more strand breaks. The P value is shown in the figure. SUPPLEMENT S1 Genes that were significantly UPREGULATED after folate intervention (by unadjusted paired t-test), list is sorted by P value Gene Symbol Nucleotide P VALUE Description OLFM4 NM_006418 0.0000 Homo sapiens differentially expressed in hematopoietic lineages (GW112) mRNA. FMR1NB NM_152578 0.0000 Homo sapiens hypothetical protein FLJ25736 (FLJ25736) mRNA. IFI6 NM_002038 0.0001 Homo sapiens interferon alpha-inducible protein (clone IFI-6-16) (G1P3) transcript variant 1 mRNA. Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 GALNTL5 NM_145292 0.0001 (GALNT15) mRNA. STIM2 NM_020860 0.0001 Homo sapiens stromal interaction molecule 2 (STIM2) mRNA. ZNF645 NM_152577 0.0002 Homo sapiens hypothetical protein FLJ25735 (FLJ25735) mRNA. ATP12A NM_001676 0.0002 Homo sapiens ATPase H+/K+ transporting nongastric alpha polypeptide (ATP12A) mRNA. U1SNRNPBP NM_007020 0.0003 Homo sapiens U1-snRNP binding protein homolog (U1SNRNPBP) transcript variant 1 mRNA. RNF125 NM_017831 0.0004 Homo sapiens ring finger protein 125 (RNF125) mRNA. FMNL1 NM_005892 0.0004 Homo sapiens formin-like (FMNL) mRNA. ISG15 NM_005101 0.0005 Homo sapiens interferon alpha-inducible protein (clone IFI-15K) (G1P2) mRNA. SLC6A14 NM_007231 0.0005 Homo sapiens solute carrier family 6 (neurotransmitter transporter) member 14 (SLC6A14) mRNA. -

Overexpressed HSF1 Cancer Signature Genes Cluster in Human Chromosome 8Q Christopher Q
Zhang et al. Human Genomics (2017) 11:35 DOI 10.1186/s40246-017-0131-5 PRIMARY RESEARCH Open Access Overexpressed HSF1 cancer signature genes cluster in human chromosome 8q Christopher Q. Zhang1,4, Heinric Williams3,4, Thomas L. Prince3,4† and Eric S. Ho1,2*† Abstract Background: HSF1 (heat shock factor 1) is a transcription factor that is found to facilitate malignant cancer development and proliferation. In cancer cells, HSF1 mediates a set of genes distinct from heat shock that contributes to malignancy. This set of genes is known as the HSF1 Cancer Signature genes or simply HSF1-CanSig genes. HSF1-CanSig genes function and operate differently than typical cancer-causing genes, yet it is involved in fundamental oncogenic processes. Results: By utilizing expression data from 9241 cancer patients, we identified that human chromosome 8q21-24 is a location hotspot for the most frequently overexpressed HSF1-CanSig genes. Intriguingly, the strength of the HSF1 cancer program correlates with the number of overexpressed HSF1-CanSig genes in 8q, illuminating the essential role of HSF1 in mediating gene expression in different cancers. Chromosome 8q21-24 is found under selective pressure in preserving gene order as it exhibits strong synteny among human, mouse, rat, and bovine, although the biological significance remains unknown. Statistical modeling, hierarchical clustering, and gene ontology-based pathway analyses indicate crosstalk between HSF1-mediated responses and pre-mRNA 3′ processing in cancers. Conclusions: Our results confirm the unique role of chromosome 8q mediated by the master regulator HSF1 in cancer cases. Additionally, this study highlights the connection between cellular processes triggered by HSF1 and pre-mRNA 3′ processing in cancers.