Evolution of Green Plants As Deduced from 5S Rrna Sequences
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

RI Equisetopsida and Lycopodiopsida.Indd
IIntroductionntroduction byby FFrancisrancis UnderwoodUnderwood Rhode Island Equisetopsida, Lycopodiopsida and Isoetopsida Special Th anks to the following for giving permission for the use their images. Robbin Moran New York Botanical Garden George Yatskievych and Ann Larson Missouri Botanical Garden Jan De Laet, plantsystematics.org Th is pdf is a companion publication to Rhode Island Equisetopsida, Lycopodiopsida & Isoetopsida at among-ri-wildfl owers.org Th e Elfi n Press 2016 Introduction Formerly known as fern allies, Horsetails, Club-mosses, Fir-mosses, Spike-mosses and Quillworts are plants that have an alternate generation life-cycle similar to ferns, having both sporophyte and gametophyte stages. Equisetopsida Horsetails date from the Devonian period (416 to 359 million years ago) in earth’s history where they were trees up to 110 feet in height and helped to form the coal deposits of the Carboniferous period. Only one genus has survived to modern times (Equisetum). Horsetails Horsetails (Equisetum) have jointed stems with whorls of thin narrow leaves. In the sporophyte stage, they have a sterile and fertile form. Th ey produce only one type of spore. While the gametophytes produced from the spores appear to be plentiful, the successful reproduction of the sporophyte form is low with most Horsetails reproducing vegetatively. Lycopodiopsida Lycopodiopsida includes the clubmosses (Dendrolycopodium, Diphasiastrum, Lycopodiella, Lycopodium , Spinulum) and Fir-mosses (Huperzia) Clubmosses Clubmosses are evergreen plants that produce only microspores that develop into a gametophyte capable of producing both sperm and egg cells. Club-mosses can produce the spores either in leaf axils or at the top of their stems. Th e spore capsules form in a cone-like structures (strobili) at the top of the plants. -

The Vascular Plants of Massachusetts
The Vascular Plants of Massachusetts: The Vascular Plants of Massachusetts: A County Checklist • First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Somers Bruce Sorrie and Paul Connolly, Bryan Cullina, Melissa Dow Revision • First A County Checklist Plants of Massachusetts: Vascular The A County Checklist First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Massachusetts Natural Heritage & Endangered Species Program Massachusetts Division of Fisheries and Wildlife Natural Heritage & Endangered Species Program The Natural Heritage & Endangered Species Program (NHESP), part of the Massachusetts Division of Fisheries and Wildlife, is one of the programs forming the Natural Heritage network. NHESP is responsible for the conservation and protection of hundreds of species that are not hunted, fished, trapped, or commercially harvested in the state. The Program's highest priority is protecting the 176 species of vertebrate and invertebrate animals and 259 species of native plants that are officially listed as Endangered, Threatened or of Special Concern in Massachusetts. Endangered species conservation in Massachusetts depends on you! A major source of funding for the protection of rare and endangered species comes from voluntary donations on state income tax forms. Contributions go to the Natural Heritage & Endangered Species Fund, which provides a portion of the operating budget for the Natural Heritage & Endangered Species Program. NHESP protects rare species through biological inventory, -

Seed Plant Models
Review Tansley insight Why we need more non-seed plant models Author for correspondence: Stefan A. Rensing1,2 Stefan A. Rensing 1 2 Tel: +49 6421 28 21940 Faculty of Biology, University of Marburg, Karl-von-Frisch-Str. 8, 35043 Marburg, Germany; BIOSS Biological Signalling Studies, Email: stefan.rensing@biologie. University of Freiburg, Sch€anzlestraße 18, 79104 Freiburg, Germany uni-marburg.de Received: 30 October 2016 Accepted: 18 December 2016 Contents Summary 1 V. What do we need? 4 I. Introduction 1 VI. Conclusions 5 II. Evo-devo: inference of how plants evolved 2 Acknowledgements 5 III. We need more diversity 2 References 5 IV. Genomes are necessary, but not sufficient 3 Summary New Phytologist (2017) Out of a hundred sequenced and published land plant genomes, four are not of flowering plants. doi: 10.1111/nph.14464 This severely skewed taxonomic sampling hinders our comprehension of land plant evolution at large. Moreover, most genetically accessible model species are flowering plants as well. If we are Key words: Charophyta, evolution, fern, to gain a deeper understanding of how plants evolved and still evolve, and which of their hornwort, liverwort, moss, Streptophyta. developmental patterns are ancestral or derived, we need to study a more diverse set of plants. Here, I thus argue that we need to sequence genomes of so far neglected lineages, and that we need to develop more non-seed plant model species. revealed much, the exact branching order and evolution of the I. Introduction nonbilaterian lineages is still disputed (Lanna, 2015). Research on animals has for a long time relied on a number of The first (small) plant genome to be sequenced was of THE traditional model organisms, such as mouse, fruit fly, zebrafish or model plant, the weed Arabidopsis thaliana (c. -

Effects of Lycopodium Clavatum and Equisetum Arvense Extracts from Western Romania
Romanian Biotechnological Letters Vol. , No. x, Copyright © 2016 University of Bucharest Printed in Romania. All rights reserved ORIGINAL PAPER Effects of Lycopodium clavatum and equisetum arvense extracts from western Romania Received for publication, July, 07, 2014 Accepted, October, 13, 2015 MARIA SUCIU1, FELIX AUREL MIC1, LUCIAN BARBU-TUDORAN2, VASILE MUNTEAN2, ALEXANDRA TEODORA GRUIA3,* 1University of Medicine and Pharmacy “Victor Babes”, Department of Functional Sciences, Timisoara, 2, Eftimie Murgu Sq., Timisoara, 300041, Timis County, Romania 2Babes-Bolyai University, Biology and Geology Department, 5-7 Clinicilor Str., Cluj-Napoca, 400084, Cluj County, Romania. 3Emergency Clinical County Hospital Timisoara, Regional Centre for Transplant Immunology Department, 10, Iosif Bulbuca Blvd., Timisoara, 300736, Timis County, Romania. *Address for correspondence to: [email protected], 10, Iosif Bulbuca Blvd., Timisoara, 300736, Timis County, Romania. Abbreviations: ALT–alanin transaminases, AST–aspartate transaminases, GC-MS–gas chromatograph coupled with mass spectrometry. Abstract Plants have always excited interest because of their active principles that could be a source of healing in various affections. The aim of this study was to demonstrate that the hepatoprotective and antimicrobial effects of Lycopodium clavatum and Equisetum arvense from the Western parts of Romania (Arad County) are not as pronounced as described in literature, against xenobiotic intoxication or microbial infection. To identify the plants active compounds, -

263 Determining the Amount of Nucleic Acids in Medicinal
Analele Universităţii din Oradea, Fascicula Protecţia Mediului Vol. XXIII, 2014 DETERMINING THE AMOUNT OF NUCLEIC ACIDS IN MEDICINAL FERNS COLLECTED FROM DIFFERENT AREAS OF ARAD AND BIHOR COUNTY Pallag Annamaria*, Osser Gyongyi**, Orodan Maria**, Honiges Ana**, Gîtea Daniela*, Pașca Bianca* *University of Oradea, Faculty of Medicine and Pharmacy, Pharmacy Department, P-ta 1 Decembrie, no. 10, Oradea, 410223, Romania, [email protected] **Vasile Goldis Western University of Arad, Romania, College of Medicine, Pharmacy and Dentistry, Biophysics Department Abstract Lycopodium clavatum L. and Equisetum arvense L. are ferns, Pteridophytes, and are widely used medicinal plants. These species are not cultivated in Romania, the medicinal product coming from traditional collection centers, so the exact origin of the product is difficult to follow. In this work we are presenting the obtained results of determining the amount of nucleic acids in case of several Lycopodium clavatum and Equisetum arvense L. populations, in terms of the species’ homogeneity. There were studied the stems of Lycopodium clavatum L. and Equisetum arvense L., collected from different regions - Oradea’s area, Cefa area - of Bihor County, and from Macea Botanical Garden (MBG), Arad county in the period of April - September 2014. As a result of the study we have observed the presence of differences in the amount of nucleic acids between the studied populations. Keywords: Lycopodium clavatum L., Equisetum arvense L., nucleic acids INTRODUCTION Pteridophytes are known from as far back as the Silurian, or some 380 million years ago (Bennici, 2008, Gifford et al, 1988). The Pteridophytes occupy the intermediate position between the bryophytes and the phanerogams (Graham, 1993). -
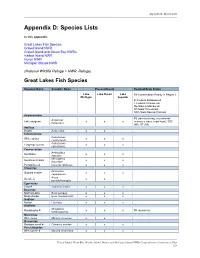
Species Lists
Appendix D: Species Lists Appendix D: Species Lists In this appendix: Great Lakes Fish Species Gravel Island NWR Gravel Island and Green Bay NWRs Harbor Island NWR Huron NWR Michigan Islands NWR (National Wildlife Refuge = NWR, Refuge) Great Lakes Fish Species Common Name Scientific Name Present/Absent Regional/State Status Lake Lake Huron Lake R3-Conservation Priority in Region 3 Michigan Superior E- Federal Endangered T-Federal Threatened SE-State Endangered ST-State Threatened SSC-State Special Concern Acipenseridae R3 (rare/declining, recreational/ Acipenser Lake sturgeon x x x economic value, tribal trust), SSC fulvescens (WI), ST (MI) Amiidae Bowfin Amia calva x x Catostomidae Catostomus White sucker x x x commersoni Catostomus Longnose sucker x x x catostomus Centrarchidae Ambloplites Rockbass x x x rupestris Micropterus Smallmouth bass x x x dolomieui Pumpkinseed Lepomis gibbosus x x x Clupeidae Dorosoma Gizzard shad # x x x cepedianum Alosa Alewife # x x pseudoharengus Cyprinidae Carp # Cyprinus Carpio x x x Esocidae Northern pike Esox Lucieus x x x Muskellunge Esox masquinongy x x x Gadidae Burbot Lota lota x x x Gobiidae Neogobius Round goby # x x x R3 (nuisance) melanostomus Moronidae White bass Morone chrysops x x Osmeridae Rainbow smelt # Osmerus mordax x x x Percichthyidae White perch # Morone americana x x x Gravel Island, Green Bay, Harbor Island, Huron, and Michigan Islands NWRs/Comprehensive Conservation Plan 221 Appendix D: Species Lists Common Name Scientific Name Present/Absent Regional/State Status Percidae R3 (rare/declining, -

Ecology and Distribution of Lycopodiaceae Mirbel in Malaysia
Blumea 54, 2009: 269–271 www.ingentaconnect.com/content/nhn/blumea RESEARCH ARTICLE doi:10.3767/000651909X476265 Ecology and distribution of Lycopodiaceae Mirbel in Malaysia G. Rusea1, K. Claysius1, S. Runi1,2, U. Joanes2, K.M. Haja Maideen3, A. Latiff 3 Key words Abstract This paper is the first account to discuss the distribution, ecology and habitats of the Lycopodiaceae in Malaysia. Lycopodiaceae are widely distributed throughout Malaysia with respect to altitudes and environmental distribution conditions but most abundantly found in hill forest and lower montane forest, terrestrial as well as epiphytic, in ecology shaded or semi-shaded places with relatively high humidity. Pahang in Peninsular Malaysia and Sabah in Borneo habitats have the highest species diversity in terms of the number of species collected. Lycopodiaceae Published on 30 October 2009 INTRODUCTION RESULTS AND DISCUSSION The Lycopodiaceae s.l. are an ancient (Correll 1956) and In Malaysia, the family comprises 32 species including 11 va- probably monophyletic family without close living relatives rieties that are found in various altitudes and vegetation types, and have a virtually cosmopolitan distribution (Øllgaard 1992). sometimes in a restricted area (Table 1). The estimated number of species ranges from approximately Lycopodiella cernua has the widest distribution in Malaysia 300 to more than 400 around the world (Wikström 2001). It and is most common on acid soils and occurs along forest consists of three genera namely Huperzia, Lycopodium and fringes, along roadside, hillsides and mountain slopes fol- Lycopodiella. Worldwide the estimated number of species for lowed by Huperzia carinata and H. pinifolia, which occur on both Lycopodium and Lycopodiella is about 40 (Wikström & tree branches. -
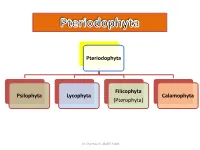
Pteriodophyta Psilophyta Lycophyta Filicophyta (Pterophyta) Calamophyta
Pteriodophyta Filicophyta Psilophyta Lycophyta Calamophyta (Pterophyta) Dr. Shaimaa N. Abd El-Fatah Class(2): Lycophyta Subclass(1): Homospora (Eligulatae) Order: Lycopodiales Family: Lycopodiaceae e.g. Lycopodium Subclass(2): Heterospora (Ligulatae) Order: Selaginellales Family: Selaginellaceae e.g. Selaginella Dr. Shaimaa N. Abd El-Fatah Phylum: Lepidophyta • It is characterized by: 1. The plant is differentiated into stem, leaves and roots. 2. Leaves are microphyllous (small, one vein and with no leaf gap). 3. The stele is protostele, siphonostele or polystele. 4. Protoxylem is exarch (ranging from a complete external cylinder to a polyarch stele in which there are many protoxylem ribs). 5. Sporangia are solitary, carried on special leaves (sporophylls). Sporophylls are usually collected in strobili. Dr. Shaimaa N. Abd El-Fatah . Among this phylum there are two independent evolutionary lines Heterospora (Ligulatae) Homospora (Eligulatae) • Characterized by presence • Eligulate (ligule lacking). of ligule (small outgrowth on • Homosporous (one type of the upper surface of the spores). leaf). • Heterosporous character (2 types of spores: microspores-- small, give rise to male gametophyte. & megaspores– larger, give rise to female gametophyte.) Dr. Shaimaa N. Abd El-Fatah • The phylum includes one class: Lycopodinae. • The class is classified into 4 orders: • Plants belong to the first order are homosporae (eligulatae), while the remaining three are heterosporae (ligulatae). Dr. Shaimaa N. Abd El-Fatah Class(2): Lycophyta Subclass(1): Homospora Subclass(2): Heterospora (Eligulatae) (Ligulatae) Order: Lycopodiales Order: Selaginellales Family: Lycopodiaceae Family: Selaginellaceae e.g. Lycopodium e.g. Selaginella Dr. Shaimaa N. Abd El-Fatah Lycopodiales This order is characterized by: 1. Homosporous and eligulatae. 2. Herbaceous without secondary growth. -

Evolution of Land Plants P
Chapter 4. The evolutionary classification of land plants The evolutionary classification of land plants Land plants evolved from a group of green algae, possibly as early as 500–600 million years ago. Their closest living relatives in the algal realm are a group of freshwater algae known as stoneworts or Charophyta. According to the fossil record, the charophytes' growth form has changed little since the divergence of lineages, so we know that early land plants evolved from a branched, filamentous alga dwelling in shallow fresh water, perhaps at the edge of seasonally-desiccating pools. The biggest challenge that early land plants had to face ca. 500 million years ago was surviving in dry, non-submerged environments. Algae extract nutrients and light from the water that surrounds them. Those few algae that anchor themselves to the bottom of the waterbody do so to prevent being carried away by currents, but do not extract resources from the underlying substrate. Nutrients such as nitrogen and phosphorus, together with CO2 and sunlight, are all taken by the algae from the surrounding waters. Land plants, in contrast, must extract nutrients from the ground and capture CO2 and sunlight from the atmosphere. The first terrestrial plants were very similar to modern mosses and liverworts, in a group called Bryophytes (from Greek bryos=moss, and phyton=plants; hence “moss-like plants”). They possessed little root-like hairs called rhizoids, which collected nutrients from the ground. Like their algal ancestors, they could not withstand prolonged desiccation and restricted their life cycle to shaded, damp habitats, or, in some cases, evolved the ability to completely dry-out, putting their metabolism on hold and reviving when more water arrived, as in the modern “resurrection plants” (Selaginella). -

Conservation Assessment for Groundcedar and Stiff Clubmoss In
United States Department of Agriculture Conservation Assessment Forest Service for Groundcedar and Stiff Rocky Mountain Region Clubmoss in the Black Black Hills National Forest Hills National Forest South Custer, South Dakota Dakota and Wyoming March 2003 J.Hope Hornbeck, Deanna J. Reyher, Carolyn Sieg and Reed W. Crook Species Assessment of Groundcedar and Stiff Clubmoss in the Black Hills National Forest, South Dakota and Wyoming J. Hope Hornbeck, Deanna J. Reyher, Carolyn Hull Sieg and Reed W. Crook J. Hope Hornbeck is a Botanist with the Black Hills National Forest in Custer, South Dakota. She completed a B.S. in Environmental Biology at The University of Montana and a M.S. in Plant Biology at the University of Minnesota. Deanna J. Reyher is an Ecologist/Soil Scientist with the Black Hills National Forest in Custer, South Dakota. She completed a B.S. degree in Agronomy from the University of Nebraska. Carolyn Hull Sieg is a Research Plant Ecologist with the Rocky Mountain Research Station in Flagstaff, Arizona. She completed a B.S. in Wildlife Biology and M.S. in Range Science from Colorado State University and a Ph.D. in Range and Wildlife Management at Texas Tech University. Reed W. Crook is a Botanist with the Black Hills National Forest in Custer, South Dakota. He completed a B.S. in Botany at Brigham Young University, and a M.S. in Plant Morphology and Ph.D. in Plant Systematics at the University of Georgia. EXECUTIVE SUMMARY Stiff clubmoss (Lycopodium annotinum L.) and groundcedar (Lycopodium complanatum L.; synonym = Diphasiastrum complanatum [L.] Holub.) (Lycopodiaceae) are circumboreal clubmoss species that are widely distributed in North American boreal habitats. -

Lycopodiaceae Clubmoss Family
Lycopodiaceae Page | 46 clubmoss family Upwards of 15 genera comprise this ancient family. Perennial herbs, they somewhat resemble coarse mosses. The solitary sporangia are borne either in a terminal strobilus or are axillary with leaves. Spores are of equal size. In Nova Scotia we have four genera. A. Rhizomes absent; upright stems clustered; axillary sporangia; spores pitted. Huperzia aa. Rhizomes present; upright shoots alternate; sporangia aggregated into B terminal strobili, spores with netlike pattern. B. Strobili on leafy peduncles; mainly of wetland habitats. Lycopodiella bb. Strobili sessile or on peduncles with remote scant leaves; mainly of C dry upland places. C. Tips of stems 5–12mm in diameter; leaves in 6 ranks or Lycopodium more; leaves bristly, free for most of their length, not scalelike. cc. Distal shoots 2–6mm in diameter; leaves in 4–6 ranks, Diphasiastrum strongly overlapping (scalelike) and appressed along the stem with only tips free. Diphasiastrum Holub There are 15–20 species worldwide; numerous hybrids are possible. Generally these clubmosses are northern or subarctic in distribution. Nova Scotia has four species. Rhizomes bear sparse leaves that are reduced to scales, rooting from the lower surfaces. Upright stems are flattened or angled, with 2–5 branches. Leaves are arranged in four ranks and of two sizes. Sporophylls are smaller than unspecialized leaves. 1-7 Lycopodiaceae Key to species A. Plants < 12 cm tall; strobili sessile. Diphasiastrum sitchense Page | 47 aa. Stems 8–50cm; strobili on peduncles. B B. Branches square or angled, bluish. D. tristachyum bb. Branches flat; green. C C. Lateral branches irregular, annual winter bud constrictions D. -

A Review of the Late Jurassic–Early Cretaceous Charophytes from The
A review of the Late Jurassic–Early Cretaceous charophytes from the northern Aquitaine Basin in south-west France Roch-Alexandre Benoit, Didier Néraudeau, Carles Martin-Closas To cite this version: Roch-Alexandre Benoit, Didier Néraudeau, Carles Martin-Closas. A review of the Late Jurassic– Early Cretaceous charophytes from the northern Aquitaine Basin in south-west France. Cretaceous Research, Elsevier, 2017, 79, pp.199-213. <10.1016/j.cretres.2017.07.009>. <insu-01574653> HAL Id: insu-01574653 https://hal-insu.archives-ouvertes.fr/insu-01574653 Submitted on 16 Aug 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Accepted Manuscript A review of the Late Jurassic–Early Cretaceous charophytes from the northern Aquitaine Basin in south-west France Roch-Alexandre Benoit, Didier Neraudeau, Carles Martín-Closas PII: S0195-6671(17)30121-0 DOI: 10.1016/j.cretres.2017.07.009 Reference: YCRES 3658 To appear in: Cretaceous Research Received Date: 13 March 2017 Revised Date: 5 July 2017 Accepted Date: 17 July 2017 Please cite this article as: Benoit, R.-A., Neraudeau, D., Martín-Closas, C., A review of the Late Jurassic–Early Cretaceous charophytes from the northern Aquitaine Basin in south-west France, Cretaceous Research (2017), doi: 10.1016/j.cretres.2017.07.009.