Novel Lncrna OXCT1-AS1 Indicates Poor Prognosis and Contributes To
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Turkish Patient with Succinyl-Coa:3-Oxoacid Coa
Original Article Journal of Inborn Errors of Metabolism & Screening 2016, Volume 4: 1–5 A Turkish Patient With ª The Author(s) 2016 DOI: 10.1177/2326409816651281 Succinyl-CoA:3-Oxoacid CoA iem.sagepub.com Transferase Deficiency Mimicking Diabetic Ketoacidosis Sahin Erdol, MD1, Mehmet Ture, MD2, Tahsin Yakut, PhD2, Halil Saglam, PhD1, Hideo Sasai, MD3, Elsayed Abdelkreem, MD3, Hiroki Otsuka, MD3, and Toshiyuki Fukao, MD, PhD3 Abstract Succinyl-CoA:3-oxoacid CoA transferase (SCOT) deficiency is an autosomal recessive disorder of ketone body utilization that is clinically characterized with intermittent ketoacidosis crises. We report here the second Turkish case with SCOT deficiency. She experienced 3 ketoacidotic episodes: The first ketoacidotic crisis mimicked diabetic ketoacidosis because of the associated hyperglycemia. Among patients with SCOT deficiency, the blood glucose levels at the first crises were variable, and this case had the highest ever reported blood glucose level. She is a compound heterozygote with 2 novel mutations, c.517A>G (K173E) and c.1543A>G (M515V), in exons 5 and 17 of the OXCT1 gene, respectively. In patient’s fibroblasts, SCOT activity was deficient and, by immunoblot analysis, SCOT protein was much reduced. The patient attained normal development and had no permanent ketosis. The accurate diagnosis of SCOT deficiency in this case had a vital impact on the management strategy and outcome. Keywords succinyl-CoA:3-oxoacid CoA transferase, deficiency, mimicking, diabetic ketoacidosis, Turkey Introduction deficiency. We herein describe a Turkish SCOT-deficient patient whose first episode mimicked diabetic ketoacidosis Succinyl-CoA:3-oxoacid CoA transferase (SCOT) deficiency because of the associated hyperglycemia. -

Novel Compound Heterozygous OXCT1 Mutations Causing Succinyl-Coa:3-Ketoacid Coa Transferase Deficiency
Case Report Yonsei Med J 2019 Mar;60(3):308-311 https://doi.org/10.3349/ymj.2019.60.3.308 pISSN: 0513-5796 · eISSN: 1976-2437 A Rare Cause of Life-Threatening Ketoacidosis: Novel Compound Heterozygous OXCT1 Mutations Causing Succinyl-CoA:3-Ketoacid CoA Transferase Deficiency Young A Kim1,2, Seong Heon Kim1, Chong Kun Cheon1, and Yoo-Mi Kim3 1Department of Pediatrics, Pusan National University Children’s Hospital, Yangsan; 2Research Institute for Convergence of Biomedical Science and Technology, Pusan National University Yangsan Hospital, Yangsan; 3Department of Pediatrics, Chungnam National University Hospital, College of Medicine, Chungnam National University, Daejeon, Korea. Succinyl-CoA:3-ketoacid CoA transferase (SCOT) deficiency is a rare inborn error of ketone body utilization, characterized by ep- isodic or permanent ketosis. SCOT deficiency is caused by mutations in the OXCT1 gene, which is mapped to 5p13 and consists of 17 exons. A 12-month-old girl presented with severe ketoacidosis and was treated with continuous renal replacement therapy. She had two previously unrecognized mild-form episodes of ketoacidosis followed by febrile illness. While high levels of ketone bodies were found in her blood and urine, other laboratory investigations, including serum glucose, were unremarkable. We identified novel compound heterozygous mutations in OXCT1:c.1118T>G (p.Ile373Ser) and a large deletion ranging from exon 8 to 16 through targeted exome sequencing and microarray analysis. This is the first Korean case of SCOT deficiency caused by novel mutations in OXCT1, resulting in life-threatening ketoacidosis. In patients with unexplained episodic ketosis, or high anion gap metabolic acidosis in infancy, an inherited disorder in ketone body metabolism should be suspected. -
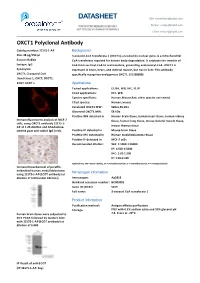
DATASHEET USA: [email protected]
DATASHEET USA: [email protected] FOR IN VITRO RESEARCH USE ONLY Europe: [email protected] NOT FOR USE IN HUMANS OR ANIMALS China: [email protected] OXCT1 Polyclonal Anbody Catalog number: 12175-1-AP Background Size: 38 μg/150 μl 3-oxoacid-CoA transferase 1 (OXCT1), encoded by nuclear gene, is a mitochondrial Source: Rabbit CoA transferase required for ketone body degradaon. It catalyzes the transfer of Isotype: IgG CoA from succinyl-CoA to acetoacetate, generang acetoacetyl-CoA. OXCT1 is Synonyms: expressed in brain, heart, and skeletal muscle, but not in liver. This anbody OXCT1; 3 oxoacid CoA specifically recognizes endogenous OXCT1. (21209089) transferase 1, OXCT, OXCT1, SCOT, SCOT s Applications Tested applicaons: ELISA, WB, IHC, IF, IP Cited applicaons: IHC, WB Species specificity: Human,Mouse,Rat; other species not tested. Cited species: Human, mouse Caculated OXCT1 MW: 520aa,56 kDa Observed OXCT1 MW: 56 kDa Posive WB detected in Human brain ssue, human heart ssue, human kidney Immunofluorescent analysis of MCF-7 ssue, human lung ssue, mouse skeletal muscle ssue, cells, using OXCT1 anbody 12175-1- AP at 1:25 diluon and Rhodamine- mouse thymus ssue labeled goat an-rabbit IgG (red). Posive IP detected in Mouse brain ssue Posive IHC detected in Human medulloblastoma ssue Posive IF detected in MCF-7 cells Recommended diluon: WB: 1:1000-1:10000 IP: 1:500-1:5000 IHC: 1:20-1:200 IF: 1:10-1:100 Applicaon key: WB = Western blong, IHC = Immunohistochemistry, IF = Immunofluorescence, IP = Immunoprecipitaon Immunohistochemical of paraffin- embedded human medulloblastoma Immunogen information using 12175-1-AP(SCOT anbody) at diluon of 1:50 (under 10x lens) Immunogen: Ag2818 GenBank accession number: BC009001 Gene ID (NCBI): 5019 Full name: 3-oxoacid CoA transferase 1 Product information Purificaon method: Angen affinity purificaon Storage: PBS with 0.1% sodium azide and 50% glycerol pH o human brain ssue were subjected to 7.3. -

Anti-OXCT1 Antibody (ARG58973)
Product datasheet [email protected] ARG58973 Package: 100 μl anti-OXCT1 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes OXCT1 Tested Reactivity Hu, Ms, Rat Tested Application IHC-P, WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name OXCT1 Antigen Species Human Immunogen Recombinant fusion protein corresponding to aa. 261-520 of Human OXCT1 (NP_000427.1). Conjugation Un-conjugated Alternate Names OXCT; EC 2.8.3.5; Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial; SCOT-s; SCOT; Somatic-type succinyl-CoA:3-oxoacid CoA-transferase; 3-oxoacid CoA-transferase 1 Application Instructions Application table Application Dilution IHC-P 1:50 - 1:100 WB 1:500 - 1:2000 Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control Mouse heart Calculated Mw 56 kDa Observed Size 56 kDa Properties Form Liquid Purification Affinity purified. Buffer PBS (pH 7.3), 0.02% Sodium azide and 50% Glycerol. Preservative 0.02% Sodium azide Stabilizer 50% Glycerol Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. www.arigobio.com 1/2 Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Gene Symbol OXCT1 Gene Full Name 3-oxoacid CoA transferase 1 Background This gene encodes a member of the 3-oxoacid CoA-transferase gene family. -

Succinyl-Coa:3-Ketoacid Coa Transferase Deficiency
Succinyl-CoA:3-ketoacid CoA transferase deficiency Description Succinyl-CoA:3-ketoacid CoA transferase (SCOT) deficiency is an inherited disorder that impairs the body's ability to break down ketones, which are molecules produced in the liver during the breakdown of fats. The signs and symptoms of SCOT deficiency typically appear within the first few years of life. Affected individuals experience episodes of extreme tiredness (lethargy), appetite loss, vomiting, rapid breathing, and, occasionally, seizures. These episodes, which are called ketoacidotic attacks, sometimes lead to coma. About half of affected individuals have a ketoacidotic attack within the first 4 days of life. Affected individuals have no symptoms of the disorder between ketoacidotic attacks. People with SCOT deficiency usually have a permanently elevated level of ketones in their blood (persistent ketosis). If the level of ketones gets too high, which can be brought on by infections, fevers, or periods without food (fasting), a ketoacidotic attack can occur. The frequency of ketoacidotic attacks varies among affected individuals. Frequency The prevalence of SCOT deficiency is unknown. More than 20 cases of this condition have been reported in the scientific literature. Causes Mutations in the OXCT1 gene cause SCOT deficiency. The OXCT1 gene provides instructions for making an enzyme called succinyl-CoA:3-ketoacid CoA transferase ( SCOT). The SCOT enzyme is made in the energy-producing centers of cells ( mitochondria). The enzyme plays a role in the breakdown of ketones, which are an important source of energy during fasting or when energy demands are increased, such as during illness or when exercising. OXCT1 gene mutations result in the production of a SCOT enzyme with little or no function. -

OXCT1 Antibody Cat
OXCT1 Antibody Cat. No.: 30-414 OXCT1 Antibody Antibody used in WB on Hum. Adult Placenta at 1 ug/ml. Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human, Mouse, Rat Antibody produced in rabbits immunized with a synthetic peptide corresponding a region IMMUNOGEN: of human OXCT1. TESTED APPLICATIONS: ELISA, WB OXCT1 antibody can be used for detection of OXCT1 by ELISA at 1:62500. OXCT1 antibody APPLICATIONS: can be used for detection of OXCT1 by western blot at 0.5 μg/mL, and HRP conjugated secondary antibody should be diluted 1:50,000 - 100,000. POSITIVE CONTROL: 1) Cat. No. 1201 - HeLa Cell Lysate PREDICTED MOLECULAR 52 kDa WEIGHT: October 2, 2021 1 https://www.prosci-inc.com/oxct1-antibody-30-414.html Properties PURIFICATION: Antibody is purified by peptide affinity chromatography method. CLONALITY: Polyclonal CONJUGATE: Unconjugated PHYSICAL STATE: Liquid Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% BUFFER: sucrose. CONCENTRATION: batch dependent For short periods of storage (days) store at 4˚C. For longer periods of storage, store STORAGE CONDITIONS: OXCT1 antibody at -20˚C. As with any antibody avoid repeat freeze-thaw cycles. Additional Info OFFICIAL SYMBOL: OXCT1 ALTERNATE NAMES: OXCT1, OXCT, SCOT ACCESSION NO.: NP_000427 PROTEIN GI NO.: 4557817 GENE ID: 5019 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References OXCT1 is a member of the 3-oxoacid CoA-transferase gene family. It is a homodimeric mitochondrial matrix enzyme that plays a central role in extrahepatic ketone body catabolism by catalyzing the reversible transfer of coenzyme A from succinyl-CoA to acetoacetate.This gene encodes a member of the 3-oxoacid CoA-transferase gene family. -

OXCT1 (NM 000436) Human Recombinant Protein – TP303764
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TP303764 OXCT1 (NM_000436) Human Recombinant Protein Product data: Product Type: Recombinant Proteins Description: Recombinant protein of human 3-oxoacid CoA transferase 1 (OXCT1), nuclear gene encoding mitochondrial protein Species: Human Expression Host: HEK293T Tag: C-Myc/DDK Predicted MW: 52 kDa Concentration: >50 ug/mL as determined by microplate BCA method Purity: > 80% as determined by SDS-PAGE and Coomassie blue staining Buffer: 25 mM Tris.HCl, pH 7.3, 100 mM glycine, 10% glycerol Preparation: Recombinant protein was captured through anti-DDK affinity column followed by conventional chromatography steps. Storage: Store at -80°C. Stability: Stable for 12 months from the date of receipt of the product under proper storage and handling conditions. Avoid repeated freeze-thaw cycles. RefSeq: NP_000427 Locus ID: 5019 UniProt ID: P55809, A0A024R040 RefSeq Size: 3572 Cytogenetics: 5p13.1 RefSeq ORF: 1560 Synonyms: OXCT; SCOT Summary: This gene encodes a member of the 3-oxoacid CoA-transferase gene family. The encoded protein is a homodimeric mitochondrial matrix enzyme that plays a central role in extrahepatic ketone body catabolism by catalyzing the reversible transfer of coenzyme A from succinyl-CoA to acetoacetate. Mutations in this gene are associated with succinyl CoA:3- oxoacid CoA transferase deficiency. [provided by RefSeq, Jul 2008] This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 OXCT1 (NM_000436) Human Recombinant Protein – TP303764 Protein Pathways: Butanoate metabolism, Synthesis and degradation of ketone bodies, Valine, leucine and isoleucine degradation Product images: Coomassie blue staining of purified OXCT1 protein (Cat# TP303764). -
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name 3-oxoacid CoA transferase 2 Gene Symbol OXCT2 Organism Human Gene Summary OXCT2 is a testis-specific succinyl-CoA:3-oxoacid CoA transferase (EC 2.8.3.5) which catalyzes the reversible transfer of CoA from succinyl-CoA to acetoacetate in the first step of ketone body utilization. See also OXCT1 (MIM 601424). Gene Aliases FLJ00030, SCOTT RefSeq Accession No. NC_000001.10, NT_032977.9 UniGene ID Hs.472491 Ensembl Gene ID ENSG00000198754 Entrez Gene ID 64064 Assay Information Unique Assay ID qHsaCED0048361 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000327582 Amplicon Context Sequence TTTTAATTAAAGTCGCAGCTCTCTAGAGGAGCACTGTCCACCTAGGGAGGAAAGT AGGAGGCCAGTAGCAAACCCCTCCCAGAGCTGGGGACA Amplicon Length (bp) 63 Chromosome Location 1:40235211-40235303 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 96 R2 0.9992 cDNA Cq 25.61 cDNA Tm (Celsius) 80.5 gDNA Cq 25.05 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report OXCT2, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. -

Increased Glucose Availability Attenuates Myocardial Ketone Body Utilization
Journal of the American Heart Association ORIGINAL RESEARCH Increased Glucose Availability Attenuates Myocardial Ketone Body Utilization Manoja K. Brahma , PhD; Chae-Myeong Ha , PhD; Mark E. Pepin , PhD; Sobuj Mia, PhD; Zhihuan Sun, BSc; John C. Chatham, PhD; Kirk M. Habegger, PhD; Evan Dale Abel , MBBS, Dphil; Andrew J. Paterson, PhD; Martin E. Young, PhD; Adam R. Wende , PhD BACKGROUND: Perturbations in myocardial substrate utilization have been proposed to contribute to the pathogenesis of car- diac dysfunction in diabetic subjects. The failing heart in nondiabetics tends to decrease reliance on fatty acid and glucose oxidation, and increases reliance on ketone body oxidation. In contrast, little is known regarding the mechanisms mediating this shift among all 3 substrates in diabetes mellitus. Therefore, we tested the hypothesis that changes in myocardial glucose utilization directly influence ketone body catabolism. METHODS AND RESULTS: We examined ventricular-cardiac tissue from the following murine models: (1) streptozotocin-induced type 1 diabetes mellitus; (2) high-fat-diet–induced glucose intolerance; and transgenic inducible cardiac-restricted expres- sion of (3) glucose transporter 4 (transgenic inducible cardiac restricted expression of glucose transporter 4); or (4) dominant negative O-GlcNAcase. Elevated blood glucose (type 1 diabetes mellitus and high-fat diet mice) was associated with reduced cardiac expression of β-hydroxybutyrate-dehydrogenase and succinyl-CoA:3-oxoacid CoA transferase. Increased myocardial β-hydroxybutyrate levels were also observed in type 1 diabetes mellitus mice, suggesting a mismatch between ketone body availability and utilization. Increased cellular glucose delivery in transgenic inducible cardiac restricted expression of glucose transporter 4 mice attenuated cardiac expression of both Bdh1 and Oxct1 and reduced rates of myocardial BDH1 activity and β-hydroxybutyrate oxidation. -

1587609685 699 2.Pdf
Life Sciences 232 (2019) 116592 Contents lists available at ScienceDirect Life Sciences journal homepage: www.elsevier.com/locate/lifescie Review article The recent insights into the function of ACAT1: A possible anti-cancer therapeutic target T Afsaneh Goudarzi Department of Clinical Biochemistry, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran ARTICLE INFO ABSTRACT Keywords: Acetoacetyl-CoA thiolase also known as acetyl-CoA acetyltransferase (ACAT) corresponds to two enzymes, one Acetyl-CoA acetyltransferase (ACAT) cytosolic (ACAT2) and one mitochondrial (ACAT1), which is thought to catalyse reversible formation of acet- Ketogenesis oacetyl-CoA from two molecules of acetyl-CoA during ketogenesis and ketolysis respectively. In addition to this Ketolysis activity, ACAT1 is also involved in isoleucine degradation pathway. Deficiency of ACAT1 is an inherited me- Post-translational modifications tabolic disorder, which results from a defect in mitochondrial acetoacetyl-CoA thiolase activity and is clinically characterized with patients presenting ketoacidosis. In this review I discuss the recent findings, which un- expectedly expand the known functions of ACAT1, indicating a role for ACAT1 well beyond its classical activity. Indeed ACAT1 has recently been shown to possess an acetyltransferase activity capable of specifically acetylating Pyruvate DeHydrogenase (PDH), an enzyme involved in producing acetyl-CoA. ACAT1-dependent acetylation of PDH was shown to negatively regulate this enzyme with a consequence in Warburg effect and tumor growth. Finally, the elevated ACAT1 enzyme activity in diverse human cancer cell lines was recently reported. These important novel findings on ACAT1's function and expression in cancer cell proliferation point to ACAT1 as a potential new anti-cancer target. -

Regulation of Autophagy by Acetyl Coenzime a : from the Mechanisms to a Revised Definition of Caloric Restriction Mimetics Federico Pietrocola
Regulation of Autophagy by Acetyl Coenzime A : From the Mechanisms to a Revised Definition of Caloric Restriction Mimetics Federico Pietrocola To cite this version: Federico Pietrocola. Regulation of Autophagy by Acetyl Coenzime A : From the Mechanisms to a Revised Definition of Caloric Restriction Mimetics. Cellular Biology. Université Paris Sud -ParisXI, 2015. English. NNT : 2015PA11T039. tel-01580849 HAL Id: tel-01580849 https://tel.archives-ouvertes.fr/tel-01580849 Submitted on 3 Sep 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. UNIVERSITE PARIS SUD ÉCOLE DOCTORALE 418 DE CANCÉROLOGIE Laboratoire : UMR1138 Apoptosis, Cancer & Immunity THESE Présentée et soutenue publiquement par FEDERICO PIETROCOLA Le 2 septembre 2015 REGULATION OF AUTOPHAGY BY ACETYL COENZIME A: FROM THE MECHANISMS TO A REVISED DEFINITION OF CALORIC RESTRICTION MIMETICS Composition du jury Directeur de thèse Pr. Guido KROEMER Directeur de Recherche UMR1138 (CRC Paris) Rapporteur Pr. Pascal FERRE Directeur CRC (CRC Paris) Dr. Patricia BOYA Directeur de Laboratoire (CIB-CSIC- Madrid) Examinateur Pr. Jose M. FUENTES Directeur de Laboratoire (UNEX RODRIGUEZ Caceres) Pr. Laurence ZITVOGEL Directeur de Recherche UMR1015 (IGR Villejuif) Dr. Maria Chiara MAIURI Chargé de Recherche UMR1138 (CRC Paris) INDEX ABBREVIATIONS ....................................................................................................................................... -

EGL Test Description
2460 Mountain Industrial Boulevard | Tucker, Georgia 30084 Phone: 470-378-2200 or 855-831-7447 | Fax: 470-378-2250 eglgenetics.com Succinyl-CoA, 3-Oxoacid CoA Transferase (SCOT) Deficiency: OXCT1 Gene Sequencing Test Code: SOXCT Turnaround time: 6 weeks CPT Codes: 81479 x1 Condition Description Succinyl-CoA:3-oxoacid CoA transferase (SCOT) deficiency causes episodic ketoacidosis without symptoms between episodes. Persistent ketoacidosis and ketonuria are characteristic of the disease, but may be absent in some affected individuals. Almost half of individuals with SCOT deficiency will develop the first ketoacidotic crisis at the age of 2-4 days. Affected individuals are general physically and developmentally normal. No diagnostic metabolites are observed in the blood and urine sample from SCOT-deficient individuals, in contrast to most organic acidemias. Ketone bodies, acetoacetate, and 3-hydroxybutyrate are, however, elevated. SCOT deficiency is caused by mutations in the OXCT1 gene (5p14). It is inherited in an autosomal recessive manner. SCOT is a mitochondrial homodimer essential for ketone body utilization. Ketone bodies, which are produced in the liver, are an important source of energy for extrahepatic tissues. References: OMIM #245050 SCOT Deficiency. OMIM #601424 OXCT1. Fukao et al. (2010). J Inherit Metab Dis. PMID: 20652411. Genes OXCT1 Indications This test is indicated for: Confirmation of a clinical diagnosis of SCOT deficiency. Carrier testing in adults with a family history of SCOT deficiency. Methodology Next Generation Sequencing: In-solution hybridization of all coding exons is performed on the patient’s genomic DNA. Although some deep intronic regions may also be analyzed, this assay is not mean to interrogate most promoter regions, deep intronic regions, or other regulatory elements, and does not detect single or multi-exon deletions or duplications.