Product Datasheet: OAAB02345
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human CD64 / FCGR1A Protein (His Tag), Biotinylated
Human CD64 / FCGR1A Protein (His Tag), Biotinylated Catalog Number: 10256-H08S-B General Information SDS-PAGE: Gene Name Synonym: CD64; Fc gamma RI Protein Construction: A DNA sequence encoding the human FCGR1A (NP_000557.1) (Met1- Pro288) was expressed with a polyhistidine tag at the C-terminus. The purified protein was biotinylated in vitro. Source: Human Expression Host: CHO Stable Cells QC Testing Purity: > 95 % as determined by SDS-PAGE. Bio Activity: Protein Description Measured by its binding ability in a functional ELISA.Immobilized High affinity immunoglobulin gamma Fc receptor I, also known as FCGR1 biotinylated Human CD64 Protein (Cat:10256-H08S-B)at 10 μg/mL can and CD64, is an integral membraneglycoprotein and a member of the bind human IgG1,The EC50 of human IgG1 is 6-14 ng/mL. immunoglobulin superfamily. CD64 is a high affinity receptor for the Fc region of IgG gamma and functions in both innate and adaptive immune Endotoxin: responses. Receptors that recognize the Fc portion of IgG function in the regulation of immune response and are divided into three classes < 1.0 EU per μg protein as determined by the LAL method. designated CD64, CD32, and CD16. CD64 is structurally composed of asignal peptidethat allows its transport to the surface of a cell, Stability: threeextracellularimmunoglobulin domainsof the C2-type that it uses to Samples are stable for up to twelve months from date of receipt at -70 ℃ bind antibody, a hydrophobictransmembrane domain, and a short cytoplasmic tail. CD64 isconstitutivelyfound on only macrophages and Predicted N terminal: Gln 16 monocytes, but treatment of polymorphonuclear leukocyteswith cytokines likeIFNγandG-CSFcan induce CD64 expression on these cells. -

FCGR1A Recombinant Protein
FCGR1A Recombinant Protein CATALOG NUMBER: 96-305 The purity of rh CD64 / FCGR1A was determined by DTT-reduced (+) SDS- PAGE and staining overnight with Coomassie Blue. Specifications SPECIES: Human SOURCE SPECIES: HEK293 cells SEQUENCE: Gln 16 - Pro 288 FUSION TAG: His Tag TESTED APPLICATIONS: WB APPLICATIONS: This recombinant protein can be used for WB. For research use only. BIOLOGICAL ACTIVITY: Measured by its ability to bind human IgG1 in the SPR assay (Biacore 2000) with the KD < 5 nM. Measured by its binding ability in a functional ELISA. Immobilized Human IgG4 at 10ug/mL (100 µl/well),can bind Human Fc gamma RI, His Tag (Cat# FCA-H52H2) with a linear of 0.1-2 ng/mL. Properties PURITY: >90% as determined by SDS-PAGE. PREDICTED MOLECULAR 32.5 kDa WEIGHT: PHYSICAL STATE: Lyopholized BUFFER: PBS, pH7.4 STORAGE CONDITIONS: Lyophilized Protein should be stored at -20˚C or lower for long term storage. Upon reconstitution, working aliquots should be stored at -20˚C or -70˚C. Avoid repeated freeze-thaw cycles. Additional Info ALTERNATE NAMES: FCGR1A, FCG1, FCGR1, IGFR1, CD64, CD64A, FCRI ACCESSION NO.: AAH32634 Background Receptors that recognize the Fc portion of IgG are divided into three groups designated Fc gamma RI, RII, and RIII, also known respectively as CD64, CD32, and CD16. Fc gamma RI binds IgG with high affinity and functions during early immune responses. Fc gamma RII and RIII are low affinity receptors that recognize IgG as aggregates surrounding multivalent antigens during late immune responses. High affinity immunoglobulin gamma Fc receptor I is also known as FCGR1A, FCG1, FCGR1, CD64 and IGFR1, is a type of integral membrane glycoprotein that binds monomeric IgG-type antibodies with high affinity, which belongs to the immunoglobulin superfamily or FCGR1 family. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Folate Receptor Β Regulates Integrin Cd11b/CD18 Adhesion of a Macrophage Subset to Collagen
Folate Receptor β Regulates Integrin CD11b/CD18 Adhesion of a Macrophage Subset to Collagen This information is current as Christian Machacek, Verena Supper, Vladimir Leksa, Goran of September 24, 2021. Mitulovic, Andreas Spittler, Karel Drbal, Miloslav Suchanek, Anna Ohradanova-Repic and Hannes Stockinger J Immunol 2016; 197:2229-2238; Prepublished online 17 August 2016; doi: 10.4049/jimmunol.1501878 Downloaded from http://www.jimmunol.org/content/197/6/2229 Supplementary http://www.jimmunol.org/content/suppl/2016/08/17/jimmunol.150187 Material 8.DCSupplemental http://www.jimmunol.org/ References This article cites 49 articles, 23 of which you can access for free at: http://www.jimmunol.org/content/197/6/2229.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on September 24, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Folate Receptor b Regulates Integrin CD11b/CD18 Adhesion of a Macrophage Subset to Collagen Christian Machacek,* Verena Supper,* Vladimir Leksa,*,† Goran Mitulovic,‡ Andreas Spittler,x Karel Drbal,{,1 Miloslav Suchanek,{ Anna Ohradanova-Repic,* and Hannes Stockinger* Folate, also known as vitamin B9, is necessary for essential cellular functions such as DNA synthesis, repair, and methylation. -

Gene List HTG Edgeseq Immuno-Oncology Assay
Gene List HTG EdgeSeq Immuno-Oncology Assay Adhesion ADGRE5 CLEC4A CLEC7A IBSP ICAM4 ITGA5 ITGB1 L1CAM MBL2 SELE ALCAM CLEC4C DST ICAM1 ITGA1 ITGA6 ITGB2 LGALS1 MUC1 SVIL CDH1 CLEC5A EPCAM ICAM2 ITGA2 ITGAL ITGB3 LGALS3 NCAM1 THBS1 CDH5 CLEC6A FN1 ICAM3 ITGA4 ITGAM ITGB4 LGALS9 PVR THY1 Apoptosis APAF1 BCL2 BID CARD11 CASP10 CASP8 FADD NOD1 SSX1 TP53 TRAF3 BCL10 BCL2L1 BIRC5 CASP1 CASP3 DDX58 NLRP3 NOD2 TIMP1 TRAF2 TRAF6 B-Cell Function BLNK BTLA CD22 CD79A FAS FCER2 IKBKG PAX5 SLAMF1 SLAMF7 SPN BTK CD19 CD24 EBF4 FASLG IKBKB MS4A1 RAG1 SLAMF6 SPI1 Cell Cycle ABL1 ATF1 ATM BATF CCND1 CDK1 CDKN1B NCL RELA SSX1 TBX21 TP53 ABL2 ATF2 AXL BAX CCND3 CDKN1A EGR1 REL RELB TBK1 TIMP1 TTK Cell Signaling ADORA2A DUSP4 HES1 IGF2R LYN MAPK1 MUC1 NOTCH1 RIPK2 SMAD3 STAT5B AKT3 DUSP6 HES5 IKZF1 MAF MAPK11 MYC PIK3CD RNF4 SOCS1 STAT6 BCL6 ELK1 HEY1 IKZF2 MAP2K1 MAPK14 NFATC1 PIK3CG RORC SOCS3 SYK CEBPB EP300 HEY2 IKZF3 MAP2K2 MAPK3 NFATC3 POU2F2 RUNX1 SPINK5 TAL1 CIITA ETS1 HEYL JAK1 MAP2K4 MAPK8 NFATC4 PRKCD RUNX3 STAT1 TCF7 CREB1 FLT3 HMGB1 JAK2 MAP2K7 MAPKAPK2 NFKB1 PRKCE S100B STAT2 TYK2 CREB5 FOS HRAS JAK3 MAP3K1 MEF2C NFKB2 PTEN SEMA4D STAT3 CREBBP GATA3 IGF1R KIT MAP3K5 MTDH NFKBIA PYCARD SMAD2 STAT4 Chemokine CCL1 CCL16 CCL20 CCL25 CCL4 CCR2 CCR7 CX3CL1 CXCL12 CXCL3 CXCR1 CXCR6 CCL11 CCL17 CCL21 CCL26 CCL5 CCR3 CCR9 CX3CR1 CXCL13 CXCL5 CXCR2 MST1R CCL13 CCL18 CCL22 CCL27 CCL7 CCR4 CCRL2 CXCL1 CXCL14 CXCL6 CXCR3 PPBP CCL14 CCL19 CCL23 CCL28 CCL8 CCR5 CKLF CXCL10 CXCL16 CXCL8 CXCR4 XCL2 CCL15 CCL2 CCL24 CCL3 CCR1 CCR6 CMKLR1 CXCL11 CXCL2 CXCL9 CXCR5 -

Identi Cation of Key Survival Genes of the Tumor Microenvironment And
Identication of key survival genes of the tumor microenvironment and immune inltration in head and neck squamous cell carcinoma Min Wang Department of Otorhinolaryngology, The First Aliated Hospital of Chongqing Medical University Tao Lu Department of Otorhinolaryngology, The First Aliated Hospital of Chongqing Medical University Yanshi Li Department of Otorhinolaryngology, The First Aliated Hospital of Chongqing Medical University Min Pan Department of Otorhinolaryngology, The First Aliated Hospital of Chongqing Medical University Zhihai Wang Department of Otorhinolaryngology, The First Aliated Hospital of Chongqing Medical University Guohua Hu ( [email protected] ) Department of Otorhinolaryngology, The First Aliated Hospital of Chongqing Medical University Research Article Keywords: Head and neck cancer, Tumor microenvironment, Immune scores, Bioinformatics analysis, Survival Posted Date: March 13th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-283947/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/23 Abstract Background: Head and neck cancer (HNC) are highly aggressive solid tumors with poor prognoses. The tumor microenvironment (TME) plays a critical role in angiogenesis, invasion, and metastasis of HNC. In the TME, immune and stromal cells inuence tumor initiation, response, and therapy. Our study aimed to evaluate the progression and prognosis of HNC by analyzing the key genes involved in immunization and stromal cells. Methods: Gene expression proles, demographics, and survival data were downloaded from the TCGA database. Patients with HNC were divided into high immune/stromal score groupss or low immune/stromal score groups based on the ESTIMATE algorithm. Differentially expressed genes (DEGs) were identied via functional enrichment analysis and protein-protein interaction networks, and survival analysis based on DEGs was also performed. -

Table S3. RAE Analysis of Well-Differentiated Liposarcoma
Table S3. RAE analysis of well-differentiated liposarcoma Model Chromosome Region start Region end Size q value freqX0* # genes Genes Amp 1 145009467 145122002 112536 0.097 21.8 2 PRKAB2,PDIA3P Amp 1 145224467 146188434 963968 0.029 23.6 10 CHD1L,BCL9,ACP6,GJA5,GJA8,GPR89B,GPR89C,PDZK1P1,RP11-94I2.2,NBPF11 Amp 1 147475854 148412469 936616 0.034 23.6 20 PPIAL4A,FCGR1A,HIST2H2BF,HIST2H3D,HIST2H2AA4,HIST2H2AA3,HIST2H3A,HIST2H3C,HIST2H4B,HIST2H4A,HIST2H2BE, HIST2H2AC,HIST2H2AB,BOLA1,SV2A,SF3B4,MTMR11,OTUD7B,VPS45,PLEKHO1 Amp 1 148582896 153398462 4815567 1.5E-05 49.1 152 PRPF3,RPRD2,TARS2,ECM1,ADAMTSL4,MCL1,ENSA,GOLPH3L,HORMAD1,CTSS,CTSK,ARNT,SETDB1,LASS2,ANXA9, FAM63A,PRUNE,BNIPL,C1orf56,CDC42SE1,MLLT11,GABPB2,SEMA6C,TNFAIP8L2,LYSMD1,SCNM1,TMOD4,VPS72, PIP5K1A,PSMD4,ZNF687,PI4KB,RFX5,SELENBP1,PSMB4,POGZ,CGN,TUFT1,SNX27,TNRC4,MRPL9,OAZ3,TDRKH,LINGO4, RORC,THEM5,THEM4,S100A10,S100A11,TCHHL1,TCHH,RPTN,HRNR,FLG,FLG2,CRNN,LCE5A,CRCT1,LCE3E,LCE3D,LCE3C,LCE3B, LCE3A,LCE2D,LCE2C,LCE2B,LCE2A,LCE4A,KPRP,LCE1F,LCE1E,LCE1D,LCE1C,LCE1B,LCE1A,SMCP,IVL,SPRR4,SPRR1A,SPRR3, SPRR1B,SPRR2D,SPRR2A,SPRR2B,SPRR2E,SPRR2F,SPRR2C,SPRR2G,LELP1,LOR,PGLYRP3,PGLYRP4,S100A9,S100A12,S100A8, S100A7A,S100A7L2,S100A7,S100A6,S100A5,S100A4,S100A3,S100A2,S100A16,S100A14,S100A13,S100A1,C1orf77,SNAPIN,ILF2, NPR1,INTS3,SLC27A3,GATAD2B,DENND4B,CRTC2,SLC39A1,CREB3L4,JTB,RAB13,RPS27,NUP210L,TPM3,C1orf189,C1orf43,UBAP2L,HAX1, AQP10,ATP8B2,IL6R,SHE,TDRD10,UBE2Q1,CHRNB2,ADAR,KCNN3,PMVK,PBXIP1,PYGO2,SHC1,CKS1B,FLAD1,LENEP,ZBTB7B,DCST2, DCST1,ADAM15,EFNA4,EFNA3,EFNA1,RAG1AP1,DPM3 Amp 1 -

FCGR2A/CD32A and FCGR3A/CD16A Variants and EULAR
FCGR2A/CD32A and FCGR3A/CD16A Variants and EULAR Response to Tumor Necrosis Factor-α Blockers in Psoriatic Arthritis: A Longitudinal Study with 6 Months of Followup JULIO RAMÍREZ, JOSÉ LUIS FERNÁNDEZ-SUEIRO, RAQUEL LÓPEZ-MEJÍAS, CARLOS MONTILLA, MAITE ARIAS, CONCEPCIÓN MOLL, MERCÉ ALSINA, RAIMON SANMARTI, FRANCISCO LOZANO, and JUAN D. CAÑETE ABSTRACT. Objective. The efficacy of antibody-based biological therapies currently used in psoriatic arthritis (PsA) depends not only on their blocking effect on the targeted molecule but also on their binding affinity to genetically defined variants of cell-surface Fc-γ receptors. Our objective was to assess the potential influence of functionally relevant FCGR2A/CD32A (H131R) and FCGR3A/CD16A (V158F) genetic polymorphisms on the EULAR response to tumor necrosis factor-α (TNF-α) blocker therapy in PsA. Methods. In total 103 patients with PsA starting anti-TNF-α therapy were included. The efficacy of therapy was evaluated according to EULAR response criteria at 3 and 6 months. FCGR2A-R131H and FCGR3A-F158V polymorphisms were genotyped. Potential correlations between clinical response and the FCGR2A-R131H and FCGR3A-F158V polymorphisms were evaluated. Results. EULAR response (moderate plus good) was 85.4% at 3 months and 87.4% at 6 months, while good EULAR response was 61.2% and 62.1%, respectively. More patients with high-affinity FCGR2A genotypes (homozygous or heterozygous combinations) achieved a EULAR response at 6 months com- pared to patients with the low-affinity genotype (RR; p = 0.034, adjusted comparison error rate < 0.025). This association was due mainly to the group of patients treated with etanercept. -

UP780, a Chromone-Enriched Aloe Composition, Enhances Adipose Insulin Receptor Signaling and Decreases Liver Lipid Biosynthesis
Open Journal of Genetics, 2013, 3, 9-86 OJGen http://dx.doi.org/10.4236/ojgen.2013.32A2002 Published Online July 2013 (http://www.scirp.org/journal/ojgen/) UP780, a Chromone-Enriched Aloe Composition, Enhances Adipose Insulin Receptor Signaling and Decreases Liver Lipid Biosynthesis Julie Tseng-Crank1*, Seon-Gil Do2, Brandon Corneliusen1, Carmen Hertel1, Jennifer Homan1, Mesfin Yimam1, Jifu Zhao1, Qi Jia1 1Unigen, Seattle, USA 2Unigen Korea, Songjung-Ri, Byeongchen-Myeon, Vheonan-Si, Chungnam, South Korea Email: *[email protected] Received 12 May 2013; revised 17 June 2013; accepted 17 July 2013 Copyright © 2013 Julie Tseng-Crank et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT 1. INTRODUCTION Nutrigenomic studies were conducted to uncover the Diabetes and prediabetes have become epidemic globally. mechanism of action for the hypoglycemic and insulin In recent US surveys, 26.9% of the population aged 65 sensitizing effects of UP780. From high fat diet-induc- years and older had diabetes and 35% of those 20 years ed obesity mouse model for UP780, livers and white or older had prediabetes [1]. WHO of the United Nations adipose tissues (WAT) from groups of lean control, estimated in 2011 that 346 million people worldwide had high fat diet (HFD), and HFD treated with UP780 were diabetes, and that number was likely to double by 2030 collected for microarray study. Microarray genera- [2]. Complications of diabetes include heart disease, ted gene expression changes were applied to Ingenu- stroke, hypertension, blindness, kidney disease, neuro- ity Pathway Analysis for changes in canonical meta- pathy, amputation, and dental disease. -
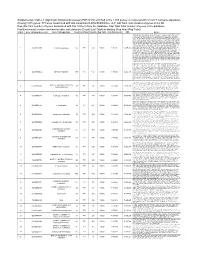
(FDR<0.05) Enriched in the 1015 Genes Co-Clustered with Known T
Supplementary Table 3. Significant biological processes (FDR<0.05) enriched in the 1,015 genes co-clustered with known T cell gene signatures. Among 1,015 genes, 771 were associated with GO annotation in DAVID database v6.7. List Total: total number of genes in my list. Pop Hits: total number of genes associated with this GO term from the database. Pop Total: total number of genes in the database. Fold Enrichment: relative enrichment ratio, calculated by (Count)/(List Total) divided by (Pop Hits)/(Pop Total). Index Gene Ontology Accession Gene Ontology Name Count List Total Pop Hits Pop Total Fold Enrichment FDR Genes AQP9, C1QC, B2M, LILRA1, LILRA2, CLEC4E, S1PR4, LILRA4, IFNG, LILRA6, CLEC4A, VNN1, ERAP2, FAS, CRTAM, C5AR1, GBP5, NCF2, NCF1, NCF4, SERPING1, HLA-DQA2, HLA-DQA1, PDCD1LG2, LILRB1, CCR9, C1QA, C1QB, LILRB2, CCR7, CCR6, UNC13D, CCR5, CD40LG, CCR4, LILRB3, CCR2, LILRB4, HLA-DPA1, VSIG4, HLA-DRA, IL1R2, IL1R1, HLA-DRB1, OAS3, ACP5, OAS1, OAS2, CD74, IFI35, ZAP70, FCER1G, HLA-DRB5, HLA-DPB1, HLA-DOA, HLA-DOB, DHX58, BLNK, IL23R, KIR2DS4, CD300C, SLAMF7, OASL, RGS1, APOL1, CD300A, HMHB1, CD209, CLEC7A, LY86, LY9, CLNK, FCRL4, SH2D1A, NOD2, HAMP, CCL3L1, CCL3L3, TICAM2, ICAM1, GZMA, CMKLR1, LY96, WAS, IL18BP, LAX1, TNFSF12- TNFSF13, HLA-DQB1, CSF2, GPR183, CCR1, GPR65, CXCL9, NCF1C, IL7R, CLEC10A, CCL24, CCL22, CYP27B1, CCL23, FCGR1C, FTHL3, FCGR1A, FCGR1B, BCL3, C2, CD27, CD28, FYB, IL18R1, IL7, CD1C, CTLA4, CCL19, CD1B, CD1A, TRIM22, CD180, CD1E, CCL18, CCL17, CCL13, FCGR2B, FCGR2C, P2RY14, LIME1, CD14, IL16, IL18, TLR1, TNFSF15, -

The Diverse Functions of the Ubiquitous Fc Receptors and Their
pathogens Review The Diverse Functions of the Ubiquitous Fcγ Receptors and Their Unique Constituent, FcRγ Subunit Thamer A. Hamdan 1,*, Philipp A. Lang 2 and Karl S. Lang 1 1 Institute of Immunology, Medical Faculty, University of Duisburg-Essen, Hufelandstraße 55, 45147 Essen, Germany 2 Department of Molecular Medicine II, Medical Faculty, Heinrich Heine University, Universitätsstrasse 1, 40225 Düsseldorf, Germany * Correspondence: [email protected] Received: 8 December 2019; Accepted: 17 February 2020; Published: 20 February 2020 Abstract: Fc gamma receptors (FcγRs) are widely expressed on a variety of immune cells and play a myriad of regulatory roles in the immune system because of their structural diversity. Apart from their indispensable role in specific binding to the Fc portion of antibody subsets, FcγRs manifest diverse biological functions upon binding to their putative ligands. Examples of such manifestation include phagocytosis, presentation of antigens, mediation of antibody-dependent cellular cytotoxicity, anaphylactic reactions, and the promotion of apoptosis of T cells and natural killer cells. Functionally, the equilibrium between activating and inhibiting FcγR maintains the balance between afferent and efferent immunity. The γ subunit of the immunoglobulin Fc receptor (FcRγ) is a key component of discrete immune receptors and Fc receptors including the FcγR family. Furthermore, FcγRs exert a key role in terms of crosslinking the innate and adaptive workhorses of immunity. Ablation of one of these receptors might positively or negatively influence the immune response. Very recently, we discovered that FcRγ derived from natural cytotoxicity triggering receptor 1 (NCR1) curtails CD8+ T cell expansion and thereby turns an acute viral infection into a chronic one. -

FCGR1A (Human) Recombinant Protein (P01)
FCGR1A (Human) Recombinant Protein (P01) Catalog # : H00002209-P01 規格 : [ 10 ug ] [ 25 ug ] List All Specification Application Image Product Human FCGR1A full-length ORF ( AAH32634.1, 1 a.a. - 374 a.a.) Enzyme-linked Immunoabsorbent Assay Description: recombinant protein with GST-tag at N-terminal. Western Blot (Recombinant Sequence: MWFLTTLLLWVPVDGQVDTTKAVITLQPPWVSVFQEETVTLHCEVLHLP protein) GSSSTQWFLNGTATQTSTPSYRITSASVNDSGEYRCQRGLSGRSDPIQ LEIHRGWLLLQVSSRVFTEGEPLALRCHAWKDKLVYNVLYYRNGKAFKF Antibody Production FHWNSNLTILKTNISHNGTYHCSGMGKHRYTSAGISVTVKELFPAPVLNAS VTSPLLEGNLVTLSCETKLLLQRPGLQLYFSFYMGSKTLRGRNTSSEYQI Protein Array LTARREDSGLYWCEAATEDGNVLKRSPELELQVLGLQLPTPVWFHVLF YLAVGIMFLVNTVLWVTIRKELKRKKKWDLEISLDSGHEKKVISSLQEDRH LEEELKCQEQKEEQLQEGVHRKEPQGAT Host: Wheat Germ (in vitro) Theoretical MW 66.77 (kDa): Preparation in vitro wheat germ expression system Method: Purification: Glutathione Sepharose 4 Fast Flow Quality Control 12.5% SDS-PAGE Stained with Coomassie Blue. Testing: Storage Buffer: 50 mM Tris-HCI, 10 mM reduced Glutathione, pH=8.0 in the elution buffer. Storage Store at -80°C. Aliquot to avoid repeated freezing and thawing. Instruction: Note: Best use within three months from the date of receipt of this protein. MSDS: Download Datasheet: Download Applications Enzyme-linked Immunoabsorbent Assay Page 1 of 3 2016/5/20 Western Blot (Recombinant protein) Antibody Production Protein Array Gene Information Entrez GeneID: 2209 GeneBank BC032634.1 Accession#: Protein AAH32634.1 Accession#: Gene Name: FCGR1A Gene Alias: CD64,CD64A,FCRI,FLJ18345,IGFR1