Supplementary Tables
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

Review Article the Role of Microbial Aspartic Protease Enzyme in Food and Beverage Industries
Hindawi Journal of Food Quality Volume 2018, Article ID 7957269, 15 pages https://doi.org/10.1155/2018/7957269 Review Article The Role of Microbial Aspartic Protease Enzyme in Food and Beverage Industries Jermen Mamo and Fassil Assefa Microbial, Cellular and Molecular Biology Department, College of Natural Science, Addis Ababa University, P.O. Box 1176, Addis Ababa, Ethiopia Correspondence should be addressed to Jermen Mamo; [email protected] Received 3 April 2018; Revised 16 May 2018; Accepted 29 May 2018; Published 3 July 2018 Academic Editor: Antimo Di Maro Copyright © 2018 Jermen Mamo and Fassil Assefa. is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Proteases represent one of the three largest groups of industrial enzymes and account for about 60% of the total global enzymes sale. According to the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology, proteases are classied in enzymes of class 3, the hydrolases, and the subclass 3.4, the peptide hydrolases or peptidase. Proteases are generally grouped into two main classes based on their site of action, that is, exopeptidases and endopeptidases. Protease has also been grouped into four classes based on their catalytic action: aspartic, cysteine, metallo, and serine proteases. However, lately, three new systems have been dened: the threonine-based proteasome system, the glutamate-glutamine system of eqolisin, and the serine-glutamate-aspartate system of sedolisin. Aspartic proteases (EC 3.4.23) are peptidases that display various activities and specicities. -

Supporting Information High-Throughput Virtual Screening
Supporting Information High-Throughput Virtual Screening of Proteins using GRID Molecular Interaction Fields Simone Sciabola, Robert V. Stanton, James E. Mills, Maria M. Flocco, Massimo Baroni, Gabriele Cruciani, Francesca Perruccio and Jonathan S. Mason Contents Table S1 S2-S21 Figure S1 S22 * To whom correspondence should be addressed: Simone Sciabola, Pfizer Research Technology Center, Cambridge, 02139 MA, USA Phone: +1-617-551-3327; Fax: +1-617-551-3117; E-mail: [email protected] S1 Table S1. Description of the 990 proteins used as decoy for the Protein Virtual Screening analysis. PDB ID Protein family Molecule Res. (Å) 1n24 ISOMERASE (+)-BORNYL DIPHOSPHATE SYNTHASE 2.3 1g4h HYDROLASE 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE 1.8 1cel HYDROLASE(O-GLYCOSYL) 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I 1.8 1vyf TRANSPORT PROTEIN 14 KDA FATTY ACID BINDING PROTEIN 1.85 1o9f PROTEIN-BINDING 14-3-3-LIKE PROTEIN C 2.7 1t1s OXIDOREDUCTASE 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE 2.4 1t1r OXIDOREDUCTASE 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE 2.3 1q0q OXIDOREDUCTASE 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE 1.9 1jcy LYASE 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE 1.9 1fww LYASE 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE 1.85 1uk7 HYDROLASE 2-HYDROXY-6-OXO-7-METHYLOCTA-2,4-DIENOATE 1.7 1v11 OXIDOREDUCTASE 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT 1.95 1x7w OXIDOREDUCTASE 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT 1.73 1d0l TRANSFERASE 35KD SOLUBLE LYTIC TRANSGLYCOSYLASE 1.97 2bt4 LYASE 3-DEHYDROQUINATE DEHYDRATASE -

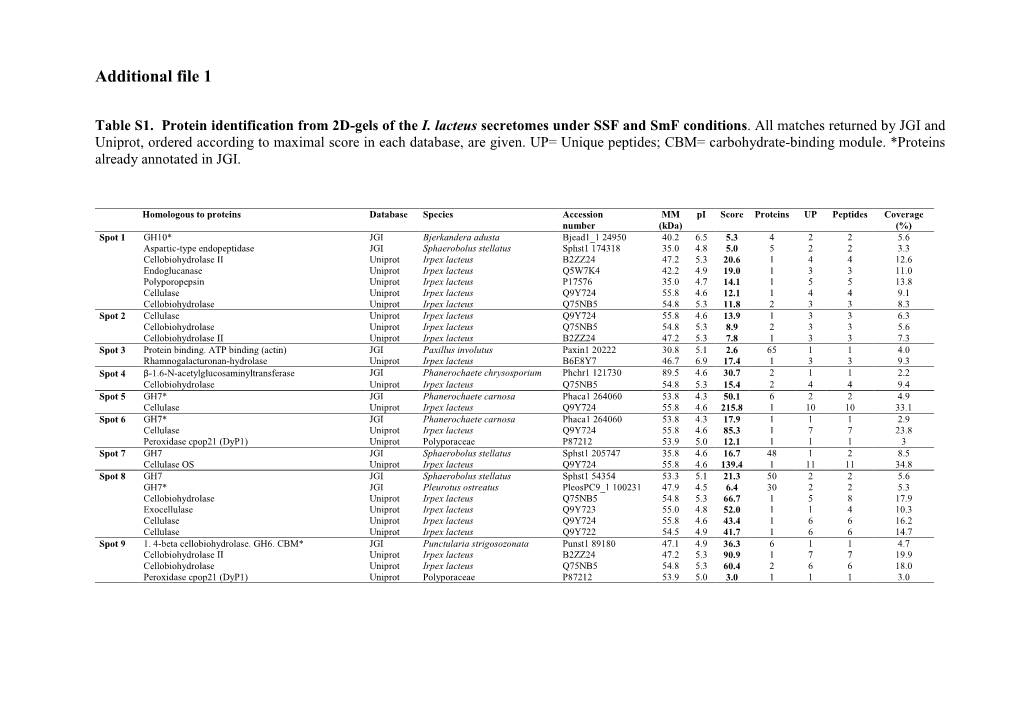
Dye-Decolorizing Peroxidases in Irpex Lacteus Combining the Catalytic
Qin et al. Biotechnol Biofuels (2018) 11:302 https://doi.org/10.1186/s13068-018-1303-9 Biotechnology for Biofuels RESEARCH Open Access Dye‑decolorizing peroxidases in Irpex lacteus combining the catalytic properties of heme peroxidases and laccase play important roles in ligninolytic system Xing Qin1,2, Huiying Luo2, Xiaoyu Zhang1, Bin Yao2, Fuying Ma1* and Xiaoyun Su2* Abstract Background: The white rot fungus Irpex lacteus exhibits a great potential in biopretreatment of lignocellulose as well as in biodegradation of xenobiotic compounds by extracellular ligninolytic enzymes. Among these enzymes, the pos- sible involvement of dye-decolorizing peroxidase (DyP) in lignin degradation is not clear yet. Results: Based on the extracellular enzyme activities and secretome analysis, I. lacteus CD2 produced DyPs as the main ligninolytic enzymes when grown in Kirk’s medium supplemented with lignin. Further transcriptome analysis revealed that induced transcription of genes encoding DyPs was accompanied by the increased expression of tran- scripts for H2O2-generating enzymes such as alcohol oxidase, pyranose 2-oxidase, and glyoxal oxidases. Meanwhile, accumulation of transcripts for glycoside hydrolase and protease was observed, in agreement with abundant pro- teins. Moreover, the biochemical analysis of IlDyP2 and IlDyP1 confrmed that DyPs were able to catalyze the oxidation of typical peroxidases substrates ABTS, phenolic lignin compounds DMP, and guaiacol as well as non-phenolic lignin compound, veratryl alcohol. More importantly, IlDyP1 enhanced catalytic activity for veratryl alcohol oxidation in the presence of mediator 1-hydroxybenzotriazole, which was similar to the laccase/1-hydroxybenzotriazole system. Conclusions: The results proved for the frst time that DyPs depolymerized lignin individually, combining catalytic features of diferent peroxidases on the functional level. -

In Silico Drug Activity Prediction of Chemical Components of Acalypha Indica Dr
International Journal of Scientific Engineering and Applied Science (IJSEAS) – Volume-2, Issue-6,June 2016 ISSN: 2395-3470 www.ijseas.com In Silico drug activity prediction of chemical components of Acalypha Indica Dr. (Mrs.)S.Shanthi M.Sc.,M.Phil.,Ph.D. Associate professor Department of chemistry,SFR college,Sivakasi. S.Sri Nisha Tharani M.Phil Chemistry ,Department of Chemistry,SFR College,Sivakasi,Tamilnadu,India. ABSTRACT Acalypha indica is a common annual Acalypha indica distributed in the herb, found mostly in the backyards of southern part of India, particularly in houses and waste places throughout the Tamilnadu has potential medicinal plains of India. Plants are used as emetic, properties and used as diuretic, anthelmintic expectorant, laxative, diuretic , bronchitis, and for respiratory problems such as pneumonia, asthma and pulmonary [1] bronchitis, asthma and pneumonia. tuberculosisP .P Leaves are laxative and Acalypha indica plant contains alkaloids, antiparasiticide; ground with common salt or tannins, steroids, saponins, terpenoids, quicklime or lime juice applied externally in flavanoids, cardiac glycosides and phenolic scabies. Leaf paste with lime juice is compounds. Some chemical components prescribed for ringworm; leaf juice is emetic were selected to theoretically evaluate their for children. A decoction of the leaves is drug likeness score using some drug given in earache. Powder of the dry leaves is designing softwares.. Their molecular given to children to expell worms; also properties were calculated using the given in the form of decoction with little software Molinspiration., Prediction of garlic. In homoepathy, the plant is used in biological activities and pharmacological severe cough associated with bleeding from activities were done using PASS online. -

Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases
Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases Alan J. Barrett Neil D. Rawlings J. Fred Woessner Editor biographies xxi Contributors xxiii Preface xxxi Introduction ' Abbreviations xxxvii ASPARTIC PEPTIDASES Introduction 1 Aspartic peptidases and their clans 3 2 Catalytic pathway of aspartic peptidases 12 Clan AA Family Al 3 Pepsin A 19 4 Pepsin B 28 5 Chymosin 29 6 Cathepsin E 33 7 Gastricsin 38 8 Cathepsin D 43 9 Napsin A 52 10 Renin 54 11 Mouse submandibular renin 62 12 Memapsin 1 64 13 Memapsin 2 66 14 Plasmepsins 70 15 Plasmepsin II 73 16 Tick heme-binding aspartic proteinase 76 17 Phytepsin 77 18 Nepenthesin 85 19 Saccharopepsin 87 20 Neurosporapepsin 90 21 Acrocylindropepsin 9 1 22 Aspergillopepsin I 92 23 Penicillopepsin 99 24 Endothiapepsin 104 25 Rhizopuspepsin 108 26 Mucorpepsin 11 1 27 Polyporopepsin 113 28 Candidapepsin 115 29 Candiparapsin 120 30 Canditropsin 123 31 Syncephapepsin 125 32 Barrierpepsin 126 33 Yapsin 1 128 34 Yapsin 2 132 35 Yapsin A 133 36 Pregnancy-associated glycoproteins 135 37 Pepsin F 137 38 Rhodotorulapepsin 139 39 Cladosporopepsin 140 40 Pycnoporopepsin 141 Family A2 and others 41 Human immunodeficiency virus 1 retropepsin 144 42 Human immunodeficiency virus 2 retropepsin 154 43 Simian immunodeficiency virus retropepsin 158 44 Equine infectious anemia virus retropepsin 160 45 Rous sarcoma virus retropepsin and avian myeloblastosis virus retropepsin 163 46 Human T-cell leukemia virus type I (HTLV-I) retropepsin 166 47 Bovine leukemia virus retropepsin 169 48 -

Supplementary Materials
Electronic Supplementary Material (ESI) for New Journal of Chemistry. This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2017 Supplementary Materials Dihydro-2H-thiopyran-3(4H)-one-1,1-dioxide - a versatile building block for the synthesis of new thiopyran-based heterocyclic systems Vitalii A. Palchykov,*a Roman M. Chabanenko,a Valeriy V. Konshin,b Victor V. Dotsenko,b,c Sergey G. Krivokolysko,b Elena A. Chigorina,d Yuriy I. Horak,e Roman Z. Lytvyn,e Andriy A. Vakhula,e Mykola D. Obushake and Alexander V. Mazepaf aDepartment of Organic Chemistry, Oles Honchar Dnipro National University, 72 Gagarina Av, 49010 Dnipro, Ukraine bDepartment of Chemistry & High Technologies, Kuban State University, 149 Stavropolskaya St, 350040 Krasnodar, Russian Federation сDepartment of Chemistry, North Caucasus Federal University, 1a Pushkin St, 355009 Stavropol, Russian Federation dFederal State Unitary Enterprise «State Scientific Research Institute of Chemical Reagents and High Purity Chemical Substances» (FSUE «IREA»), 3 Bogorodsky val St, 107076 Moscow, Russian Federation eDepartment of Organic Chemistry, Ivan Franko National University of Lviv, 6 Kyryla i Mefodiya St, 79005 Lviv, Ukraine fA. V. Bogatsky Physico-Chemical Institute, National Academy of Sciences of Ukraine, 86 Lustdorfskaya Rd, 65080 Odessa, Ukraine *E-mail: [email protected] 1 CONTENTS: Page: 1 H and 13C NMR spectra of compound 2a (400/100 MHz, DMSO-d6)................................ 4 IR spectrum of compound 2a ............................................................................................. 4 Mass spectrum (EI) of compound 2a ................................................................................. 5 1 H-1H COSY spectrum of compound 2a (400 MHz, DMSO-d6) .......................................... 5 NOESY spectrum of compound 2a (400 MHz, DMSO-d6) ................................................. 6 1 H-13C HSQC spectrum of compound 2a (400/100 MHz, DMSO-d6) ................................ -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

(12) United States Patent (10) Patent No.: US 8,561,811 B2 Bluchel Et Al
USOO8561811 B2 (12) United States Patent (10) Patent No.: US 8,561,811 B2 Bluchel et al. (45) Date of Patent: Oct. 22, 2013 (54) SUBSTRATE FOR IMMOBILIZING (56) References Cited FUNCTIONAL SUBSTANCES AND METHOD FOR PREPARING THE SAME U.S. PATENT DOCUMENTS 3,952,053 A 4, 1976 Brown, Jr. et al. (71) Applicants: Christian Gert Bluchel, Singapore 4.415,663 A 1 1/1983 Symon et al. (SG); Yanmei Wang, Singapore (SG) 4,576,928 A 3, 1986 Tani et al. 4.915,839 A 4, 1990 Marinaccio et al. (72) Inventors: Christian Gert Bluchel, Singapore 6,946,527 B2 9, 2005 Lemke et al. (SG); Yanmei Wang, Singapore (SG) FOREIGN PATENT DOCUMENTS (73) Assignee: Temasek Polytechnic, Singapore (SG) CN 101596422 A 12/2009 JP 2253813 A 10, 1990 (*) Notice: Subject to any disclaimer, the term of this JP 2258006 A 10, 1990 patent is extended or adjusted under 35 WO O2O2585 A2 1, 2002 U.S.C. 154(b) by 0 days. OTHER PUBLICATIONS (21) Appl. No.: 13/837,254 Inaternational Search Report for PCT/SG2011/000069 mailing date (22) Filed: Mar 15, 2013 of Apr. 12, 2011. Suen, Shing-Yi, et al. “Comparison of Ligand Density and Protein (65) Prior Publication Data Adsorption on Dye Affinity Membranes Using Difference Spacer Arms'. Separation Science and Technology, 35:1 (2000), pp. 69-87. US 2013/0210111A1 Aug. 15, 2013 Related U.S. Application Data Primary Examiner — Chester Barry (62) Division of application No. 13/580,055, filed as (74) Attorney, Agent, or Firm — Cantor Colburn LLP application No. -

Identification and Functional Characterization of Effectors from an Anther Smut Fungus, Microbotryum Lychnidis-Dioicae
University of Louisville ThinkIR: The University of Louisville's Institutional Repository Electronic Theses and Dissertations 12-2018 Identification and functional characterization of effectors from an anther smut fungus, Microbotryum lychnidis-dioicae. Venkata Swathi Kuppireddy University of Louisville Follow this and additional works at: https://ir.library.louisville.edu/etd Part of the Molecular Genetics Commons Recommended Citation Kuppireddy, Venkata Swathi, "Identification and functional characterization of effectors from an anther smut fungus, Microbotryum lychnidis-dioicae." (2018). Electronic Theses and Dissertations. Paper 3078. https://doi.org/10.18297/etd/3078 This Doctoral Dissertation is brought to you for free and open access by ThinkIR: The University of Louisville's Institutional Repository. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of ThinkIR: The University of Louisville's Institutional Repository. This title appears here courtesy of the author, who has retained all other copyrights. For more information, please contact [email protected]. IDENTIFICATION AND FUNCTIONAL CHARACTERIZATION OF EFFECTORS FROM AN ANTHER SMUT FUNGUS, MICROBOTRYUM LYCHNIDIS-DIOICAE By Venkata Swathi Kuppireddy M.Sc., Biology, University of Louisville, 2016 A Dissertation Submitted to the Faculty of the College of Arts and Sciences of the University of Louisville in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in Biology Department of Biology, Division of Molecular, Cellular and Developmental Biology, University of Louisville Louisville, Kentucky December 2018 Copyright 2018 by Venkata Swathi Kuppireddy © All rights reserved IDENTIFICATION AND FUNCTIONAL CHARACTERIZATION OF EFFECTORS FROM AN ANTHER SMUT FUNGUS, MICROBOTRYUM LYCHNIDIS-DIOICAE By Venkata Swathi Kuppireddy M.Sc., Biology, University of Louisville, 2016 A Dissertation Approved on November 14, 2018 By the following Dissertation Committee: _________________________________________ Dr. -

Discovery of High Affinity Receptors for Dityrosine Through Inverse Virtual Screening and Docking and Molecular Dynamics
Article Discovery of High Affinity Receptors for Dityrosine through Inverse Virtual Screening and Docking and Molecular Dynamics Fangfang Wang 1,*,†, Wei Yang 2,3,† and Xiaojun Hu 1,* 1 School of Life Science, Linyi University, Linyi 276000, China; [email protected] 2 Department of Microbiology, Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia, [email protected] 3 Arieh Warshel Institute of Computational Biology, the Chinese University of Hong Kong, 2001 Longxiang Road, Longgang District, Shenzhen 518000, China * Corresponding author: [email protected] † These authors contributed equally to this work. Received: 09 December 2018; Accepted: 23 December 2018; Published: date Table S1. Docking affinity scores for cis-dityrosine binding to binding proteins. Target name PDB/UniProtKB Type Affinity (kcal/mol) Galectin-1 1A78/P56217 Lectin -6.2±0.0 Annexin III 1AXN/P12429 Calcium/phospholipid Binding Protein -7.5±0.0 Calmodulin 1CTR/P62158 Calcium Binding Protein -5.8±0.0 Seminal Plasma Protein Pdc-109 1H8P/P02784 Phosphorylcholine Binding Protein -6.6±0.0 Annexin V 1HAK/P08758 Calcium/phospholipid Binding -7.4±0.0 Alpha 1 antitrypsin 1HP7/P01009 Protein Binding -7.6±0.0 Histidine-Binding Protein 1HSL/P0AEU0 Binding Protein -6.3±0.0 Intestinal Fatty Acid Binding Protein 1ICN/P02693 Binding Protein(fatty Acid) -9.1±0.0* Migration Inhibitory Factor-Related Protein 14 1IRJ/P06702 Metal Binding Protein -7.0±0.0 Lysine-, Arginine-, Ornithine-Binding Protein 1LST/P02911 Amino Acid Binding Protein -6.5±0.0 -

Proteomics Reveals Synergy in Biomass Conversion Between Fungal Enzymes and Inorganic Fenton
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.11.239541; this version posted August 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Proteomics reveals synergy in biomass conversion between fungal enzymes and inorganic Fenton 2 chemistry in leaf-cutting ant colonies 3 4 5 6 Morten Schiøtt*, Jacobus J. Boomsma 7 8 Centre for Social Evolution, Department of Biology, University of Copenhagen, Universitetsparken 9 15, DK-2100 Copenhagen, Denmark 10 11 12 13 Email addresses: 14 Morten Schiøtt: [email protected] 15 Jacobus J. Boomsma: [email protected] 16 17 * Corresponding author 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.11.239541; this version posted August 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 18 Abstract 19 The herbivorous symbiosis between leaf-cutting ants and fungal cultivars processes biomass via ant 20 fecal fluid mixed with munched plant substrate before fungal degradation. Here we present a full 21 proteome of the fecal fluid of Acromyrmex leaf-cutting ants, showing that most proteins function as 22 biomass degrading enzymes and that ca. 80% are produced by the fungal cultivar and ingested, but not 23 digested, by the ants. Hydrogen peroxide producing oxidoreductases were remarkably common in the 24 fecal proteome, inspiring us to test a scenario in which hydrogen peroxide reacts with iron in the fecal 25 fluid to form reactive oxygen radicals after which oxidized iron is reduced by other fecal-fluid 26 enzymes.