SUPPLEMENTARY TABLES LIST of TABLES 1 List of Primers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

2006 Catalog
Friends School of Minnesota Nonprofit Org. U.S. Postage 1365 Englewood Avenue PAID Saint Paul, MN 55104 Minneapolis, MN Permit No. 1767 TIME VALUE DATA If you have received a duplicate copy, please let us know, and pass the extra to a friend! North Star Originals 6 The Himalayan Saint Paul, Blue Poppy FROM 35W Minnesota FROM HWY 3 What’s “Native” LARPENTEUR AVENUE SNELLING AVE Mean When It Comes to Plants? Minnesota State Fair Friends School CLEVELAND AVE Plant Sale 280 COMMONWEALTH DAN PATCH MIDWAY PKWY P May 12, 13, 14, 2006 Friday,May 12 36 COMO AVENUE Cleveland 35W Snelling 11:00 A.M.–8:00 P.M. Larpenteur CANFIELD Saturday,May 13 State Fair Grandstand PLANT SALE 9:00 A.M.–8:00 P.M. Y 280 Como G PAR ER K N FROM 94 E Sunday,May 14 Friends School NOON P.M. The native Penstemon grandiflorus 12:00 –4:00 (Large-Flowered Beardtongue), 94 photographed in St. Paul’s At the State Fair Midway area during summer 2005. Grandstand 17th Annual Friends School Plant Sale May 12th, 13th and 14th, 2006 Friday 11:00 A.M.–8:00 P.M.• Saturday 9:00 A.M.–8:00 P.M. Sunday 12:00 NOON–4:00 P.M.Sunday is half-price day at the Minnesota State Fair Grandstand Friends School of Minnesota Thank you for supporting Friends School of Minnesota by purchasing plants at our sale. Friends School of Minnesota prepares children to embrace life, learning, and community with hope, skill, understanding, and creativity. We are committed to the Quaker values of peace, justice, simplicity and integrity. -

Genetic and Molecular Basis of Petal Pigmentation in Floriculture Crops: a Review
Available online at www.ijpab.com Dhakar and Soni Int. J. Pure App. Biosci. 6 (1): 442-451 (2018) ISSN: 2320 – 7051 DOI: http://dx.doi.org/10.18782/2320-7051.5192 ISSN: 2320 – 7051 Int. J. Pure App. Biosci. 6 (1): 442-451 (2018) Review Article Genetic and Molecular Basis of Petal Pigmentation in Floriculture Crops: A Review Sunita Dhakar1* and Anjali Soni2 1Division of Floriculture and Landscaping, ICAR- IARI, New Delhi 110012, India 2Division of Fruits and Horticultural Technology, ICAR-IARI, New Delhi 110012, India *Corresponding Author E-mail: [email protected] Received: 11.07.2017 | Revised: 19.08.2017 | Accepted: 26.08.2017 ABSTRACT Flower colours are of paramount importance in the ecology of plants and their ability to attract pollinators and seed dispersing organisms. The primary pigments occurring in plants are chlorophylls and carotenoids accumulated in plastids, anthocyanins and betalains, which are dissolved in vacuolar sap. Flavonoids and carotenoids are widely distributed in plant pigments. Among the factors influencing flower colour, anthocyanin biosynthesis has been the most extensively studied. Each plant species usually accumulates limited kinds of anthocyanins and exhibits limited flower colour. For example, roses and carnations lack violet/blue colour because they do not accumulate delphinidin-based anthocyanins and petunia and cymbidium lack brick red/orange colour due to the lack of pelargonidin-based anthocyanins. Different approaches are used for development of novel flower colour in floricultural crops like hybridization, mutation and genetic engineering. Advances in molecular biology and plant transformation technology have made possible the engineering of an anthocyanin biosynthetic pathway by the overexpression of heterologous flavonoid biosynthetic genes or the down-regulation of endogenous genes in transgenic plants. -

SUPPLEMENTARY TABLES LIST of TABLES 1 List of Universal and Diagnostic Primer
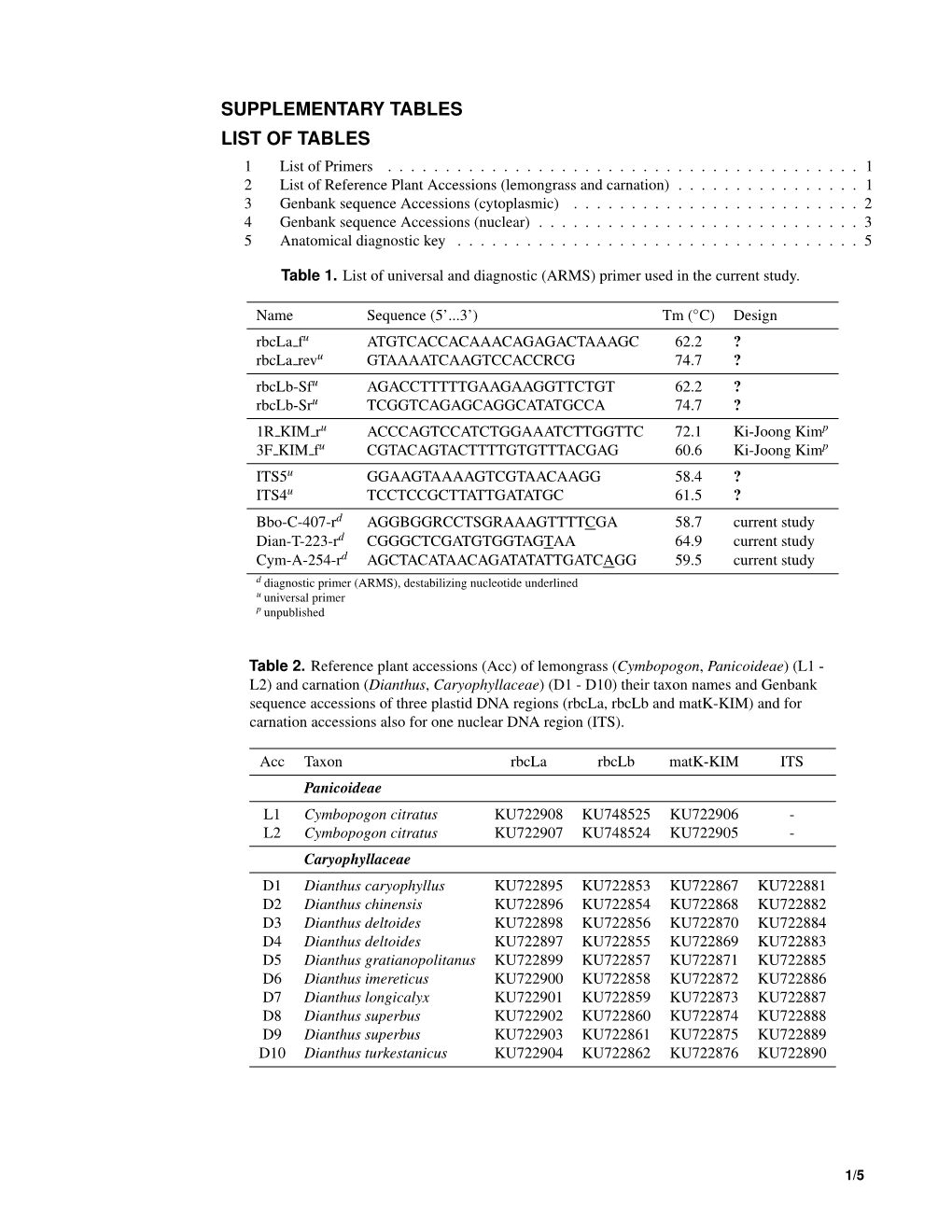
SUPPLEMENTARY TABLES LIST OF TABLES 1 List of universal and diagnostic primer . .1 3 Genbank sequence accessions (cytoplasmic) . .1 4 Genbank sequence accessions (nuclear) . .2 2 List of secondary specimens (lemongrass and carnation) . .5 5 Anatomical diagnostic key . .6 6 BLOG bamboo tribe classification results . .7 Table 1. List of universal and diagnostic (ARMS) primer used in the current study. The table includes primer names and sequences, melting temperatures (Tm) of each individual primer, the annealing temperatures (An) for each primer pair and references to corresponding publications. Name Sequence (5’...3’) Tm (◦C) An (◦C) Design rbcLa fu ATGTCACCACAAACAGAGACTAAAGC 62.2 58 Kondo et al. [1996] rbcLa revu GTAAAATCAAGTCCACCRCG 74.7 Fofana et al. [1997] rbcLb-Sfu AGACCTTTTTGAAGAAGGTTCTGT 62.2 55 Dong et al. [2014] rbcLb-Sru TCGGTCAGAGCAGGCATATGCCA 74.7 1R KIM ru ACCCAGTCCATCTGGAAATCTTGGTTC 72.1 52 Ki-Joong Kimp 3F KIM fu CGTACAGTACTTTTGTGTTTACGAG 60.6 ITS5u GGAAGTAAAAGTCGTAACAAGG 58.4 56 White et al. [1990] ITS4u TCCTCCGCTTATTGATATGC 61.5 Bbo-C-407-rd AGGBGGRCCTSGRAAAGTTTTCGA 58.7 current study Dian-T-223-rd CGGGCTCGATGTGGTAGTAA 64.9 Cym-A-254-rd AGCTACATAACAGATATATTGATCAGG 59.5 d diagnostic primer (ARMS), destabilizing nucleotide underlined u universal primer p unpublished Table 3. GenBank sequence accessions of two plastid DNA regions (rbcL and matK) used in the current study. Taxon rbcL matK Arundinarieae Arundinaria gigantea subsp. tecta AJ746179 EF125165 Borinda emeryi EF125079 EF125167 Chimonobambusa marmorea AJ746176 EF125168 Drepanostachyum falcatum AJ746265 EF125170 Fargesia dracocephala AJ746266 KP685610 Fargesia sp. Asmussen AM110249 AM114722 Indocalamus latifolius AJ746177 EF125173 Phyllostachys aurea HE573324 AF164390 Phyllostachys bambusoides AB088833 AB088805 Phyllostachys nigra HE573325 EU434241 Pleioblastus maculatus JN247242 JN247148 Pseudosasa amabilis AJ746273 KP093752 Pseudosasa japonica FN870405 HG794001 Thamnocalamus spathiflorus EF125087 KP685639 Bambubseae Bambusa multiplex M91626 EF125166 1/7 Table 3. -

Phylogeny, Ecology and Plant Features in Tropical and Mediterranean Communities
Research Interspecific variation across angiosperms in global DNA methylation: phylogeny, ecology and plant features in tropical and Mediterranean communities Conchita Alonso1 ,Monica Medrano1 , Ricardo Perez2, Azucena Canto3 ,Vıctor Parra-Tabla4 and Carlos M. Herrera1 1Estacion Biologica de Donana,~ CSIC, Avenida Americo Vespucio 26, 41092, Sevilla, Spain; 2Instituto de Investigaciones Quımicas, Centro de Investigaciones Cientıficas Isla de La Cartuja, CSIC-US, Avenida Americo Vespucio 49, 41092, Sevilla, Spain; 3Centro de Investigacion Cientıfica de Yucatan, A.C., Calle 43 No. 130 x 32 y 34, Chuburna de Hidalgo, 97205, Merida, Yucatan, Mexico; 4Departamento de Ecologıa Tropical, Universidad Autonoma de Yucatan, Campus de Ciencias Biologicas y Agropecuarias, Km. 15.5 Carretera Merida-Xtmakui, 97000, Merida, Yucatan, Mexico Summary Author for correspondence: The interspecific range of epigenetic variation and the degree to which differences between Conchita Alonso angiosperm species are related to geography, evolutionary history, ecological settings or Tel: +34 954 466 700 species-specific traits, remain essentially unexplored. Genome-wide global DNA cytosine Email: [email protected] methylation is a tractable ‘epiphenotypic’ feature suitable for exploring these relationships. Received: 16 April 2019 Global cytosine methylation was estimated in 279 species from two distant, ecologically dis- Accepted: 28 June 2019 parate geographical regions: Mediterranean Spain and tropical Mexico. At each region, four distinct plant communities were analyzed. New Phytologist (2019) Global methylation spanned a 10-fold range among species (4.8–42.2%). Interspecific dif- doi: 10.1111/nph.16046 ferences were related to evolutionary trajectories, as denoted by a strong phylogenetic signal. Genomes of tropical species were on average less methylated than those of Mediterranean Key words: DNA methylation, epigenetics, ones. -

Distribution and Diversity of Cytotypes in Dianthus Broteri As Evidenced by Genome Size Variations
Annals of Botany 104: 965–973, 2009 doi:10.1093/aob/mcp182, available online at www.aob.oxfordjournals.org Distribution and diversity of cytotypes in Dianthus broteri as evidenced by genome size variations Francisco Balao*, Ramo´n Casimiro-Soriguer, Marı´a Talavera, Javier Herrera and Salvador Talavera Departamento de Biologı´a Vegetal y Ecologı´a, Universidad de Sevilla, Apdo. 1095, E-41080 Sevilla, Spain Received: 20 April 2009 Returned for revision: 26 May 2009 Accepted: 15 June 2009 Published electronically: 25 July 2009 Downloaded from https://academic.oup.com/aob/article/104/5/965/138784 by guest on 30 September 2021 † Background and Aims Studying the spatial distribution of cytotypes and genome size in plants can provide valu- able information about the evolution of polyploid complexes. Here, the spatial distribution of cytological races and the amount of DNA in Dianthus broteri, an Iberian carnation with several ploidy levels, is investigated. † Methods Sample chromosome counts and flow cytometry (using propidium iodide) were used to determine overall genome size (2C value) and ploidy level in 244 individuals of 25 populations. Both fresh and dried samples were investigated. Differences in 2C and 1Cx values among ploidy levels within biogeographical pro- vinces were tested using ANOVA. Geographical correlations of genome size were also explored. † Key Results Extensive variation in chromosomes numbers (2n ¼ 2x ¼ 30, 2n ¼ 4x ¼ 60, 2n ¼ 6x ¼ 90 and 2n ¼ 12x ¼180) was detected, and the dodecaploid cytotype is reported for the first time in this genus. As regards cytotype distribution, six populations were diploid, 11 were tetraploid, three were hexaploid and five were dodecaploid. -

Pucciniomycotina: Microbotryum) Reflect Phylogenetic Patterns of Their Caryophyllaceous Hosts
Org Divers Evol (2013) 13:111–126 DOI 10.1007/s13127-012-0115-1 ORIGINAL ARTICLE Contrasting phylogenetic patterns of anther smuts (Pucciniomycotina: Microbotryum) reflect phylogenetic patterns of their caryophyllaceous hosts Martin Kemler & María P. Martín & M. Teresa Telleria & Angela M. Schäfer & Andrey Yurkov & Dominik Begerow Received: 29 December 2011 /Accepted: 2 October 2012 /Published online: 6 November 2012 # Gesellschaft für Biologische Systematik 2012 Abstract Anther smuts in the genus Microbotryum often is a factor that should be taken into consideration in delimitat- show very high host specificity toward their caryophyllaceous ing species. Parasites on Dianthus showed mainly an arbitrary hosts, but some of the larger host groups such as Dianthus are distribution on Dianthus hosts, whereas parasites on other crucially undersampled for these parasites so that the question Caryophyllaceae formed well-supported monophyletic clades of host specificity cannot be answered conclusively. In this that corresponded to restricted host groups. The same pattern study we sequenced the internal transcribed spacer (ITS) was observed in the Caryophyllaceae studied: morphological- region of members of the Microbotryum dianthorum species ly described Dianthus species did not correspond well with complex as well as their Dianthus hosts. We compared phy- monophyletic clades based on molecular data, whereas other logenetic trees of these parasites including sequences of anther Caryophyllaceae mainly did. We suggest that these different smuts from other Caryophyllaceae, mainly Silene,withphy- patterns primarily result from different breeding systems and logenies of Caryophyllaceae that are known to harbor anther speciation times between different host groups as well as smuts. Additionally we tested whether observed patterns in difficulties in species delimitations in the genus Dianthus. -

The Evolution of Dianthus Polylepis Complex (Caryophyllaceae) Inferred from Morphological and Nuclear DNA Sequence Data: One Or Two Species?
Plant Syst Evol (2013) 299:1419–1431 DOI 10.1007/s00606-013-0804-z ORIGINAL ARTICLE The evolution of Dianthus polylepis complex (Caryophyllaceae) inferred from morphological and nuclear DNA sequence data: one or two species? Mohammad Farsi • Maryam Behroozian • Jamil Vaezi • Mohammad Reza Joharchi • Farshid Memariani Received: 3 December 2012 / Accepted: 22 March 2013 / Published online: 6 April 2013 Ó Springer-Verlag Wien 2013 Abstract Dianthus polylepis complex consists of two Keywords Caryophyllaceae Á Dianthus polylepis already known endemic species, Dianthus polylepis and complex Á Morphology Á Molecular phylogeny Á Iran D. binaludensis, in Khorassan-Kopetdagh floristic prov- ince. The taxonomic position of these species has long been debated. The aim of the present study is to shed light Introduction on the evolutionary relationships of the members of the complex using morphological and molecular data. In One of the fundamental problems in plant taxonomy con- morphological study, firstly, 56 vegetative and floral cerns with the plant identification, especially for distin- characters were measured on 33 specimens of the both guishing closely related or recently evolved species species. Multivariate analyses were performed on 25 (out (Rieseberg et al. 2006; Fazekas et al. 2009; Yan et al. of 56) significantly discriminating morphological traits. In 2011). molecular study, we sequenced alleles obtained from a Dianthus L. (Caryophyllaceae) with over 300 species region between 2nd and 6th exons of the gene coding for worldwide is characterized by its extensive morphological the enzyme dihydroflavonol 4-reductase copy1 (DFR1). variability at both inter- and intraspecific levels (Erhardt Morphological results show that most of a priori identified 1990, 1991; Friedman et al. -

University of Montenegro Faculty of Sciences and Mathematics
UNIVERSITY OF MONTENEGRO FACULTY OF SCIENCES AND MATHEMATICS PROTECTED AREA GAP ASSESSMENT WITH COMPREHENSIVE PLAN FOR A REPRESENTATIVE PAS (PROTECTED AREA SYSTEM) November, 2012th 1 PROJETC TEAM: DR DANILO MRDAK – Project leader, expert for fish, for conservation issues and for sustainable development DR DANKA CAKOVIĆ – Expert for botany and conservation issues MR SNEŽANA VUKSANOVIĆ – Expert for botany DR MARKO KARAMAN – Expert for invertebrate fauna DR RUŽA ĆIROVIĆ – Expert for herpetofauna MR NELA VEŠOVIĆ – DUBAK – Expert for bird fauna DR VESNA MAČIĆ – Expert on marine flora and fauna MR MARINA ĐUROVIĆ - Expert on mammal fauna DR SNEŽANA DRAGIĆEVIĆ – Expert on moss DR GORDANA KASOM – Expert on fungi DR DRAGANA MILOŠEVIĆ – Expert for conservation issues MR ANA KATNIĆ – Expert for sustainable development DR SEONA ANDERSON – International expert for conservation issues DR PREDRAG STANIŠIĆ - Expert for data basis MR MARJAN ERCEG – Expert for GIS 2 3 1. IDENTIFICATION, ASSESSING AND MAPPING OF HABITATS AND SPECIES DISTRIBUTION 1.1. Identification and assessing Experts for botany reviewed list of habitats related to Montenegro (lend and marine). They paid attention on habitats that were specified as important for Emerald network designation as well as to ones that were of interest for Yearly Monitoring of Biodiversity status 2011th. The Botany team identifies habitats that are either endangered or threatened as well as the ones that are rare or specific for Balkan nature. They produce check list of habitats that serve as a referent check -

Alpine Garden Club of British Columbia Seed Exchange 2015
Alpine GArden Club of british ColumbiA seed exChAnGe 2015 We are very grateful to all those members who have made our Seed Exchange possible through donating seeds, and to those living locally who volunteer so much time and effort to packaging and filling orders. We especially appreciate those donors who made an extra effort in this difficult summer. We have fewer donors but still a good list and that is thanks to them. Read the following instructions carefully before filling out the seed request form. PLEASE KEEP YOUR SEED LIST, packets will be marked by number only. Please send the Seed Exchange Request Form (last page of booklet) by mail as soon as possible, but no later than DECEMBER 10. You can request your seeds on-line (see back page) and the complete seed list is available on the website for reference. This year you will find a special list of plants at the end of the ‘Native to North America’ section, of plants wild collected on Pink Mountain in northern BC. Please see the accompanying note in the November Bulletin. The Club would appreciate it if anyone who raises plants from these seeds and obtains seed from them would let us know. Those seeds may be useful in the future for revegetation projects. Allocation: Donors may receive up to 60 packets and non-donors 30 packets, limit of one packet of each selection. Donors receive preference for seeds in short supply. (USDA will permit no more than 50 packets for those living in the USA.) List first choices by number only, in strict numerical order, from left to right on the order form. -

Lenka Šafářová POLYPLOIDNÍ KOMPLEX ALLIUM OLERACEUM L
UNIVERZITA PALACKÉHO V OLOMOUCI PŘÍRODOV ĚDECKÁ FAKULTA KATEDRA BOTANIKY Lenka Šafá řová POLYPLOIDNÍ KOMPLEX ALLIUM OLERACEUM L. V EVROP Ě POLYPLOID COMPLEX OF ALLIUM OLERACEUM L. IN EUROPE DOKTORSKÁ DISERTA ČNÍ PRÁCE Studijní obor botanika Školitel: RNDr. Martin Duchoslav, Ph.D. Olomouc 2011 Bibliografická identifikace Jméno a p říjmení autora: Lenka Šafá řová Název práce: Polyploidní komplex Allium oleraceum L. v Evrop ě Typ práce: doktorská Pracovišt ě: Katedra botaniky – UP Olomouc, Východo české muzeum v Pardubicích Studijní program: Biologie Studijní obor: Botanika Školitel: RNDr. Martin Duchoslav, PhD. Rok obhajoby: 2011 Abstrakt: Polyploidie je p řírodní proces, hrající významnou úlohu v evoluci rostlin, většina z nich ve své minulosti pravd ěpodobn ě prod ělala genovou duplikaci následovanou procesem diploidizace. S využitím metody pr ůtokové cytometrie významn ě stoupla možnost studia polyploidie, struktury polyploidních komplex ů i cytotypové variability jednotlivých rostlinných druh ů. Jako modelový organizmus pro zkoumání polyploidie byl zvolen česnek planý (Allium oleraceum L). Jedná se o polyploidní komplex, u n ěhož byly na po čátku výzkumu známé čty ři ploidní úrovn ě: triploidní (2n=3x=24), tetraploidní (2n=4x=32), pentaploidní (2n=5x=40) a hexaploidní (2n=6x=48). Sb ěr vzork ů probíhal na t řech odlišných prostorových škálách – na úrovni populací, na úrovní p řibližného st ředu rozší ření druhu (Česká Republika, Slovensko) a na úrovni Evropy. Celkem bylo studováno 620 populací a metodou pr ůtokové cytometrie analyzováno 7354 rostlin. Byly potvrzeny všechny doposud známé ploidní úrovn ě, nov ě byly zjišt ěny heptaploidní populace (2n=7x=56) a jedenkrát byla nalezena populace oktoploidní (2n=8x=64). -

2018 Catalog Compressed.Pdf
2018 Plant Catalogue Hemerocallis ‘Bold Tiger’ Bold Tiger Daylily As bold as its name, bright orange petals with Zone 4-9 Height Spacing a large red eye and yellow-green throat. 28” 18-24” Bloom Color Petals are dramatically recurved with a nicely Zone Culture ruffled edge. Well-branched with heavily Orange w/ Red Eye Sun to Part Shade budded scapes. Tetraploid. Dormant winter foliage. Rabbit resistant. Moist, Well Drained Bloom Season Attracts Mid Summer Butterflies, Hummers Hemerocallis ‘Brookwood Black Kitten’ Brookwood Black Kitten Daylily Thus unusual color will wow you and your Zone 3-9 Height Spacing guests with its Black Red flower with 22” 18-24” Bloom Color contrasting midrib stripe. Zone Culture Diploid. Dormant winter foliage. Black Red Sun to Part Shade Rabbit resistant. Moist, Well Drained Bloom Season Attracts Mid Summer Butterflies, Hummers Hemerocallis ‘Celebrity Elite’ Celebrity Elite Daylily A cherry-red flower with yellow throats over Zone 3-9 Height Spacing grass-like leaf blades. Blooms in mid July. 25” 18-24” Bloom Color Dormant foliage in winter. Diploid. Zone Culture Red Self Sun to Part Shade Moist, Well Drained Bloom Season Attracts Mid Summer Butterflies, Hummers Hemerocallis ‘Chicago Apache’ Chicago Apache Daylily Intense scarlet red blooms with a golden Zone 4-9 Height Spacing throat are 5” across. Petals have a loosely 28” 18-24” Bloom Color ruffled edge. Very sun-fast. Zone Culture Dormant foliage, Tetraploid. Red Self Sun or Part Shade Rabbit resistant. Moist, Well Drained Bloom Season Attracts Mid-Late Summer Butterflies, Hummers Hemerocallis ‘Daring Deception’ Daring Deception Daylily Features dusky cream-pink trumpet-shaped Zone 3-9 Height Spacing flowers with a burgundy-purple throat and 24” 18-24” Bloom Color pie-crust edging. -
Serbia and Montenegro Biodiversity Analysis, 2002
FAA SECTION 119 BIODIVERSITY ANALYSIS FOR SERBIA AND MONTENEGRO Prepared for: United States Agency for International Development Mission to the Federal Republics of Yugoslavia Submitted By: Loren L. Schulze and The Environmental Information Systems and Networking Project (Contract No. EE-C-00-98-00001-00) DevTech Systems, Inc. May 2002 Final Report Note This document was prepared under the auspices of United States Agency for International Development (USAID) by a Biodiversity Analysis Team. The team was comprised of Dr. Loren L. Schulze, team leader and consultants from the Environmental Information Systems and Networking Project managed by DevTech Systems, Inc. Dr. Bruce A Byers consultant ecologist was responsible for the ecological analysis and Violeta Orlovic was responsible for the institutional analysis. The statements and opinions expressed herein are not necessarily those of either USAID or DevTech Systems, Inc. Table of Contents Table of Contents ............................................................................................................................... 1 List of Appendices ............................................................................................................................. 3 Executive Summary ........................................................................................................................... 4 Introduction ........................................................................................................................................ 5 Section One: Conservation