Aves, Emberizinae): Integrating Ecology, Evolution, and Systematics" (2006
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

North Andean Origin and Diversification of the Largest Ithomiine Butterfly Genus
North Andean origin and diversification of the largest ithomiine butterfly genus The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Lisa De-Silva, D., L. L. Mota, N. Chazot, R. Mallarino, K. L. Silva- Brandão, L. M. G. Piñerez, A. V. Freitas, et al. 2017. “North Andean origin and diversification of the largest ithomiine butterfly genus.” Scientific Reports 7 (1): 45966. doi:10.1038/srep45966. http:// dx.doi.org/10.1038/srep45966. Published Version doi:10.1038/srep45966 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:32630680 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA www.nature.com/scientificreports OPEN North Andean origin and diversification of the largest ithomiine butterfly genus Received: 31 October 2016 Donna Lisa De-Silva1, Luísa L. Mota2, Nicolas Chazot1,3, Ricardo Mallarino4, Karina L. Silva- Accepted: 22 February 2017 Brandão5, Luz Miryam Gómez Piñerez6,7, André V.L. Freitas2, Gerardo Lamas8, Published: 07 April 2017 Mathieu Joron9, James Mallet4, Carlos E. Giraldo6,10, Sandra Uribe6, Tiina Särkinen11, Sandra Knapp12, Chris D. Jiggins13, Keith R. Willmott14 & Marianne Elias1 The Neotropics harbour the most diverse flora and fauna on Earth. The Andes are a major centre of diversification and source of diversity for adjacent areas in plants and vertebrates, but studies on insects remain scarce, even though they constitute the largest fraction of terrestrial biodiversity. -

Butterflies (Lepidoptera: Papilionoidea) in a Coastal Plain Area in the State of Paraná, Brazil
62 TROP. LEPID. RES., 26(2): 62-67, 2016 LEVISKI ET AL.: Butterflies in Paraná Butterflies (Lepidoptera: Papilionoidea) in a coastal plain area in the state of Paraná, Brazil Gabriela Lourenço Leviski¹*, Luziany Queiroz-Santos¹, Ricardo Russo Siewert¹, Lucy Mila Garcia Salik¹, Mirna Martins Casagrande¹ and Olaf Hermann Hendrik Mielke¹ ¹ Laboratório de Estudos de Lepidoptera Neotropical, Departamento de Zoologia, Universidade Federal do Paraná, Caixa Postal 19.020, 81.531-980, Curitiba, Paraná, Brazil Corresponding author: E-mail: [email protected]٭ Abstract: The coastal plain environments of southern Brazil are neglected and poorly represented in Conservation Units. In view of the importance of sampling these areas, the present study conducted the first butterfly inventory of a coastal area in the state of Paraná. Samples were taken in the Floresta Estadual do Palmito, from February 2014 through January 2015, using insect nets and traps for fruit-feeding butterfly species. A total of 200 species were recorded, in the families Hesperiidae (77), Nymphalidae (73), Riodinidae (20), Lycaenidae (19), Pieridae (7) and Papilionidae (4). Particularly notable records included the rare and vulnerable Pseudotinea hemis (Schaus, 1927), representing the lowest elevation record for this species, and Temenis huebneri korallion Fruhstorfer, 1912, a new record for Paraná. These results reinforce the need to direct sampling efforts to poorly inventoried areas, to increase knowledge of the distribution and occurrence patterns of butterflies in Brazil. Key words: Atlantic Forest, Biodiversity, conservation, inventory, species richness. INTRODUCTION the importance of inventories to knowledge of the fauna and its conservation, the present study inventoried the species of Faunal inventories are important for providing knowledge butterflies of the Floresta Estadual do Palmito. -
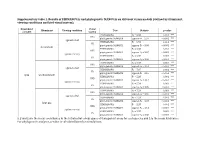
Supplementary Table 1. Results of Permanovas and Phylogenetic Manovas on Different Vision Models (Defined by Illuminant, Viewing Conditions and Bird Visual System)
Supplementary table 1. Results of PERMANOVAs and phylogenetic MANOVAs on different vision models (defined by illuminant, viewing conditions and bird visual system). Dependent Visual Illuminant Viewing condition Test Statistic p-value variable system PERMANOVA F9 = 6.88 0.001 *** UVS phylogenetic MANOVA approx-F9 = 2.97 < 0.001 *** against a leaf PERMANOVA F9 = 6.93 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.05 < 0.001 *** forest shade PERMANOVA F9 = 5.38 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.07 < 0.001 *** against the sky PERMANOVA F9 = 5.38 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.36 < 0.001 *** PERMANOVA F9 = 7.04 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.01 < 0.001 *** against a leaf PERMANOVA F9 = 7.07 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.10 < 0.001 *** xyzL woodland shade PERMANOVA F9 = 5.33 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.12 < 0.001 *** against the sky PERMANOVA F9 = 5.34 0.002 ** VS phylogenetic MANOVA approx-F9 = 3.39 < 0.001 *** PERMANOVA F9 = 7.24 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.00 < 0.001 *** against a leaf PERMANOVA F9 = 7.24 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.07 < 0.001 *** large gap PERMANOVA F9 = 5.37 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.14 < 0.001 *** against the sky PERMANOVA F9 = 5.37 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.38 < 0.001 *** x, y and z are the mean coordinates in the tetrahedral colour space of transparent areas for each species and L is the mean luminance. -

Elevation in Tropical Sky Islands As the Common Driver in Structuring Genes and Communities of Freshwater Organisms
Research Collection Journal Article Elevation in tropical sky islands as the common driver in structuring genes and communities of freshwater organisms Author(s): Gueuning, Morgan; Suchan, Tomasz; Rutschmann, Sereina; Gattolliat, Jean-Luc; Jamsari, Jamsari; Kamil, Al I.; Pitteloud, Camille; Buerki, Sven; Balke, Michael; Sartori, Michel; Alvarez, Nadir Publication Date: 2017 Permanent Link: https://doi.org/10.3929/ethz-b-000217957 Originally published in: Scientific Reports 7(1), http://doi.org/10.1038/s41598-017-16069-y Rights / License: Creative Commons Attribution 4.0 International This page was generated automatically upon download from the ETH Zurich Research Collection. For more information please consult the Terms of use. ETH Library www.nature.com/scientificreports OPEN Elevation in tropical sky islands as the common driver in structuring genes and communities of Received: 30 January 2017 Accepted: 7 November 2017 freshwater organisms Published: xx xx xxxx Morgan Gueuning1,2, Tomasz Suchan1,12, Sereina Rutschmann1,3, Jean-Luc Gattolliat1,4, Jamsari Jamsari5, Al Ihsan Kamil5, Camille Pitteloud1,6,7, Sven Buerki8,9, Michael Balke10, Michel Sartori 1,4 & Nadir Alvarez 1,11 Tropical mountains are usually characterized by a vertically-arranged sequence of ecological belts, which, in contrast to temperate habitats, have remained relatively stable in space across the Quaternary. Such long-lasting patterning of habitats makes them ideal to test the role of environmental pressure in driving ecological and evolutionary processes. Using Sumatran freshwater mayfy communities, we test whether elevation, rather than other spatial factors (i.e. volcanoes, watersheds) structures both species within communities and genes within species. Based on the analysis of 31 mayfy (Ephemeroptera) communities and restriction-site-associated-DNA sequencing in the four most ubiquitous species, we found elevation as the major spatial component structuring both species and genes in the landscape. -

Rev Iss Web Mec 13773 25-22 5765..5784
Molecular Ecology (2016) 25, 5765–5784 doi: 10.1111/mec.13773 Into the Andes: multiple independent colonizations drive montane diversity in the Neotropical clearwing butterflies Godyridina NICOLAS CHAZOT,*† KEITH R. WILLMOTT,‡ FABIEN L. CONDAMINE,§¶ DONNA LISA DE-SILVA,* ANDREV.L.FREITAS,**GERARDOLAMAS,†† HELENE MORLON,‡‡ CARLOS E. GIRALDO,§§ CHRIS D. JIGGINS,¶¶ MATHIEU JORON,*** JAMES MALLET,††† SANDRA URIBE‡‡‡ and MARIANNE ELIAS* *Institut de Systematique, Evolution, Biodiversite, ISYEB – UMR 7205 – CNRS MNHN UPMC EPHE, Museum national d’Histoire naturelle, Sorbonne Universites, 57 rue Cuvier CP50, F-75005 Paris, France, †Department of Biology, University of Lund, 223 62 Lund, Sweden, ‡McGuire Center for Lepidoptera and Biodiversity, Florida Museum of Natural History, University of Florida, Gainesville, FL 32611, USA, §CNRS, UMR 5554 Institut des Sciences de l’Evolution (Universite de Montpellier), Place Eugene Bataillon, 34095 Montpellier, France, ¶Department of Biological Sciences, University of Alberta, T6G 2E9 Edmonton, AB, Canada, **Departamento de Zoologia and Museu de Zoologia, Instituto de Biologia, Universidade Estadual de Campinas, Campinas, S~ao Paulo, Brazil, ††Museo de Historia Natural, Universidad Nacional de San Marcos, Lima, Peru, ‡‡IBENS, Ecole Normale Superieure, UMR 8197 CNRS, Paris, France, §§Grupo de Investigacion de Sanidad Vegetal, Universidad Catolica de Oriente, Rionegro, Antioquia, Colombia, ¶¶Department of Zoology, University of Cambridge, Cambridge, UK, ***Centre d’Ecologie Fonctionnelle et Evolutive, CEFE, UMR 5175 CNRS – EPHE – Universite de Montpellier – Universite Paul Valery Montpellier, 34293 Montpellier 5, France, †††Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA 02138, USA, ‡‡‡Universidad Nacional de Colombia, sede Medellın, Medellın, Colombia Abstract Understanding why species richness peaks along the Andes is a fundamental question in the study of Neotropical biodiversity. -

Molecular Phylogenetics of the Neotropical Butterfly Subtribe Oleriina
Molecular Phylogenetics and Evolution 55 (2010) 1032–1041 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Molecular phylogenetics of the neotropical butterfly subtribe Oleriina (Nymphalidae: Danainae: Ithomiini) Donna Lisa de-Silva a,*, Julia J. Day a, Marianne Elias b,c, Keith Willmott d, Alaine Whinnett a, James Mallet a a Department of Genetics, Evolution and Environment, University College London, Wolfson House, 4 Stephenson Way, London NW1 2HE, UK b Imperial College London, Silwood Park, Buckhurst Road, Ascot, Berkshire SL5 7PY, UK c CNRS, UMR 7205, Muséum National d’Histoire Naturelle, 45 Rue Buffon, CP50, 75005 Paris, France d McGuire Center for Lepidoptera, Florida Museum of Natural History, University of Florida, P.O. Box 112710, Gainesville, FL 32611-2710, USA article info abstract Article history: The Oleriina is one of the most speciose subtribes of the neotropical nymphalid butterfly tribe Ithomiini. Received 9 September 2009 They are widely distributed across the Andes and Amazonian lowlands and like other ithomiines they are Revised 22 December 2009 involved in complex mimicry rings. This subtribe is of particular interest because it contains the most Accepted 9 January 2010 diverse ithomiine genus, Oleria, as well as two genera, Megoleria and Hyposcada, that feed on hostplants Available online 15 January 2010 not utilized elsewhere in the tribe. Here we present the first comprehensive species-level phylogeny for the Oleriina, representing 83% of recognised species in the group, and based on 6698 bp from eight mito- Keywords: chondrial (mt) and nuclear (nc) genes. Topologies are largely congruent for ncDNA and the concatenated Lepidoptera dataset and the genera Oleria, Hyposcada and Megoleria are recovered and well-supported, although Speciation Phylogeny strongly discordant genealogy between mtDNA and ncDNA suggest possible introgression among Hypos- Hybridization cada and Megoleria. -

Illustrations of New Species of Exotic Butterflies
^^3 ILLUSTRATIONS OP NEW SPECIES OP EXOTIC BUTTERFLIES WILLIAM C. HBWITSON. lit SELECTED CHIEFLY FEOM HIS OWN COLLECTION. VOL. V. JOHN VAN VOORST, LONDON. 1872—1876. LONDON : PRINTED BY WOODFALL AND KINDER, . MILFORD LANE, STRAND, W.O. PREFACE. It is with regret, not however unmixed with satisfaction, tliat I come to the close of a work which has been to me a twenty-five years' labour of love. With regret that age and failing health forbid me to commence another volume. With satisfaction when I remember the great kindness which I have experienced personally from aU lepidopterists during its progress, and the very favourable reception it has met with from aE, and especially from those whose position as naturalists gives value to their opinions. I have many times during the progress of the book expressed my grati- tude for the kindness and liberality which I have met with from Dr. Boisduval, not only in giving me free access to his collections, but in allowing me during his absence to select from them and bring home with me aU that I wished to figure, feeling, as I do, the difiiculty I should myself experience in being parted from any of my treasures. To the generous encouragement met with from Mr. Wilson Saunders, especially at its commencement, the work owes much of its success. I am fully aware of and regret many errors, but have endeavoured to atone for them as much as possible by myself pointing them out and correcting them. It has always been my study to make the work useful rather than attractive. -

Curriculum Vitae
CURRICULUM VITAE Jason Piers Wilton Hall Department of Entomology National Museum of Natural History Smithsonian Institution Washington, DC, USA Tel: (202) 633 1047 Email: [email protected] Education: 1994-1999 Ph.D., Department of Entomology and Nematology, University of Florida, Gainesville, USA 1990-1993 B.A., M.A. Honours, Department of Zoology, St. Johns College, Oxford University, Oxford, UK Grants and Awards: 2007-2011 National Science Foundation Grant (Principal Investigator on Biodiversity Surveys & Inventories Grant DEB# 0639977 - “Collaborative Research: The Butterflies of Ecuador (Lepidoptera: Papilionoidea) - A Comprehensive Survey of a Megadiverse Fauna”) 2003 Royal Entomological Society Award (for best paper in the journal Systematic Entomology during 2001-2 – Willmott, K. R., J. P. W. Hall, and G. Lamas. Systematics of Hypanartia (Lepidoptera: Nymphalidae: Nymphalinae), with a test for speciation mechanisms in the Andes) 2001-2006 National Science Foundation Grant (Principal Investigator on Biodiversity Surveys & Inventories Grant # 0103746 - “Survey of the Andean butterfly fauna of Ecuador”) 2001-2002 Smithsonian Postdoctoral Fellowship (National Museum of Natural History, Washington, DC) 1999-2000 Smithsonian Postdoctoral Fellowship (National Museum of Natural History, Washington, DC) 1999 J. Turner Predoctoral Fellowship (University of Florida, Gainesville, FL) 1998 Collections Studies Grant (American Museum of Natural History, New York, NY) 1997-2000 National Geographic Society Grant (Joint Principal Investigator -

Phylogenetic Revision of Eryphanis Boisduval, with a Description of a New Species from Ecuador (Lepidoptera, Nymphalidae)
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Center for Systematic Entomology, Gainesville, Insecta Mundi Florida June 2008 Phylogenetic Revision of Eryphanis Boisduval, with a Description of a New Species from Ecuador (Lepidoptera, Nymphalidae) Carla M. Penz University of New Orleans, New Orleans, Louisiana Follow this and additional works at: https://digitalcommons.unl.edu/insectamundi Part of the Entomology Commons Penz, Carla M., "Phylogenetic Revision of Eryphanis Boisduval, with a Description of a New Species from Ecuador (Lepidoptera, Nymphalidae)" (2008). Insecta Mundi. 562. https://digitalcommons.unl.edu/insectamundi/562 This Article is brought to you for free and open access by the Center for Systematic Entomology, Gainesville, Florida at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Insecta Mundi by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. INSECTA MUNDI A Journal of World Insect Systematics 0035 Phylogenetic Revision of Eryphanis Boisduval, with a Description of a New Species from Ecuador (Lepidoptera, Nymphalidae) Carla M. Penz Department of Biological Sciences University of New Orleans 2000 Lakeshore Drive New Orleans, Louisiana, 70148, USA [email protected] Date of Issue: June 6, 2008 CENTER FOR SYSTEMATIC ENTOMOLOGY, INC., Gainesville, FL Carla M. Penz Phylogenetic Revision of Eryphanis Boisduval, with a Description of a New Species from Ecuador (Lepidoptera, Nymphalidae) Insecta Mundi 0035: 1-25 Published in 2008 by Center for Systematic Entomology, Inc. P. O. Box 147100 Gainesville, FL 32614-7100 U. S. A. http://www.centerforsystematicentomology.org/ Insecta Mundi is a journal primarily devoted to insect systematics, but articles can be published on any non-marine arthropod taxon. -

Nymphalidae: Ithomiinae)
STUDIES ON THE ECOLOGY AND EVOLUTION OF NEOTROPICAL ITHOMIINE BUTTERFLIES (NYMPHALIDAE: ITHOMIINAE) by GEORGE WILLIAM BECCALONI A thesis submitted for the degree of Doctor ofPhilosophy ofthe University ofLondon October 1995 Biogeography and Conservation Laboratory Centre for Population Biology Department of Entomology Imperial College The Natural History Museum Silwood Park Cromwell Road Ascot London SW7 5BD Berkshire SL5 7PY 2 To my mother, Benjie & Judy in love and gratitude 3 ABSTRACT Two aspects ofthe ecology ofNeotropical ithomiine butterflies (Nymphalidae: Ithomiinae) are discussed: mimicry (Chapters 2, 3) and species richness (Chapters 4, 5). Chapter 2 defines eight mimicry complexes involving ithomiines and other insects found in eastern Ecuador. These complexes are dominated by ithomiine individuals. Hypotheses to explain polymorphism in Batesian and Mullerian mimics are assessed. In Chapter 3, evidence that sympatric ithomiine-dominated mimicry complexes are segregated by microhabitat is reviewed. Data confirm that sympatric complexes are segregated vertically by flight height. Flight height is shown to be positively correlated with larval host-plant height. Host-plant partitioning between species in a butterfly community results in the formation of microhabitat guilds of species, and evidence suggests that mimicry may evolve between species which share a guild, but not between guilds. Models for the evolution of mimicry complexes in sympatry, and for polymorphism and dual sex-limited mimicry in Mullerian mimics, are discussed in the light of these findings. Chapter 4 investigates relationships between species richness offamilies and subfamilies ofNeotropical butterflies and overall butterfly species richness at local and regional scales. A strong positive correlation is demonstrated between ithomiine richness and the species richness of all other butterflies. -

Diversification of Clearwing Butterflies with the Rise of the Andes
Diversification of clearwing butterflies with the rise of the Andes The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation De#Silva, Donna Lisa, Marianne Elias, Keith Willmott, James Mallet, and Julia J. Day. 2015. “Diversification of clearwing butterflies with the rise of the Andes.” Journal of Biogeography 43 (1): 44-58. doi:10.1111/jbi.12611. http://dx.doi.org/10.1111/jbi.12611. Published Version doi:10.1111/jbi.12611 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:29002675 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Journal of Biogeography (J. Biogeogr.) (2016) 43, 44–58 ORIGINAL Diversification of clearwing butterflies ARTICLE with the rise of the Andes Donna Lisa De-Silva1,2*, Marianne Elias1, Keith Willmott3, James Mallet4 and Julia J. Day2 1Institut de Systematique, Evolution, ABSTRACT Biodiversite, ISYEB – UMR 7205 – CNRS, Aim Despite the greatest butterfly diversity on Earth occurring in the Neotrop- MNHN, UPMC, EPHE, Museum National ical Andes and Amazonia, there is still keen debate about the origins of this d’Histoire Naturelle, Sorbonne Universites, 57 rue Cuvier, CP50, Paris F-75005, France, exceptional biota. A densely sampled calibrated phylogeny for a widespread 2Department of Genetics, Evolution and butterfly subtribe, Oleriina (Nymphalidae: Ithomiini) was used to estimate the Environment, University College London, origin, colonization history and diversification of this species-rich group. -

Running Head 1 the AGE of BUTTERFLIES REVISITED
bioRxiv preprint doi: https://doi.org/10.1101/259184; this version posted February 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Running head 2 THE AGE OF BUTTERFLIES REVISITED (AND TESTED) 3 Title 4 The Trials and Tribulations of Priors and Posteriors in Bayesian Timing of 5 Divergence Analyses: the Age of Butterflies Revisited. 6 7 Authors 8 NICOLAS CHAZOT1*, NIKLAS WAHLBERG1, ANDRÉ VICTOR LUCCI FREITAS2, 9 CHARLES MITTER3, CONRAD LABANDEIRA3,4, JAE-CHEON SOHN5, RANJIT KUMAR 10 SAHOO6, NOEMY SERAPHIM7, RIENK DE JONG8, MARIA HEIKKILÄ9 11 Affiliations 12 1Department of Biology, Lunds Universitet, Sölvegatan 37, 223 62, Lund, Sweden. 13 2Departamento de Biologia Animal, Instituto de Biologia, Universidade Estadual de 14 Campinas (UNICAMP), Cidade Universitária Zeferino Vaz, Caixa postal 6109, 15 Barão Geraldo 13083-970, Campinas, SP, Brazil. 16 3Department of Entomology, University of Maryland, College Park, MD 20742, U.S.A. 17 4Department of Paleobiology, National Museum of Natural History, Smithsonian 18 Institution, Washington, DC 20013, USA; Department of Entomology and BEES 19 Program, University of Maryland, College Park, MD 20741; and Key Lab of Insect 20 Evolution and Environmental Change, School of Life Sciences, Capital Normal 21 University, Beijing 100048, bioRxiv preprint doi: https://doi.org/10.1101/259184; this version posted February 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity.