Phylogenesis of Brain-Derived Neurotrophic Factor (BDNF) In
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Cytogenetic and Genetic Evidence of Male Sexual Inversion by Heat Treatment in the Newt Pleurodeles Poireti
CHROMOSOMA Chromosoma (Bet1) (1984) 90:261-264 Springer-Verlag 1984 Cytogenetic and genetic evidence of male sexual inversion by heat treatment in the newt Pleurodeles poireti C. Dournon 1, F. Guillet 1, D. Boucher 2, and J.C. Lacroix 2 1 Laboratoire de Biologic Animale; 2Laboratoire de G6n~tique du D~veloppement, Universit~ P. et M. Curie, 9 quai St Bernard 75230 Paris Cedex 05, France Abstract. Larvae of Pleurodeles poireti were maintained the lengthy reproductive cycle of the amphibians. It should during their development at a high temperature (31 ~ C). be interesting therefore to identify the genotypic sex of an In several species of amphibians, such a treatment is known experimental animal by cytogenetic or other genetic criteria, to change the sex ratio through the inversion of genotypic i.e. through sex chromosomes or sex-linked characters re- females into phenotypic males. Pleurodeles poireti is an ex- spectively. ception. It is the first reported amphibian in which heat In a geographical race of P. poireti, the sex chromo- induces an inversion of genotypic males into functional phe- somes can be distinguished in preparations of lampbrush notypic females. The sexual genotype of standard and ex- chromosomes of the oocytes (Lacroix 1970). Moreover, perimental phenotypic females was determined through he- Ferrier et al. (1980, 1983), have shown that in P. waltlii terochromosomes in lampbrush stage. In the present study, the enzyme peptidase I shows a sex-linked polymorphism. we have utilised another technique for identification of sex- The effects of heat treatment on sexual differentiation ual genotype, applicable to both phenotypic males and fe- of larvae of P. -

Short Note Development and Characterization of Twelve New
*Manuscript Click here to download Manuscript: Isolation and characterization of microsatellite loci for Pleurodeles waltl_final_rev.docx 1 Short Note 2 Development and characterization of twelve new polymorphic microsatellite loci in 3 the Iberian Ribbed newt, Pleurodeles waltl (Caudata: Salamandridae), with data 4 on cross-amplification in P. nebulosus 5 JORGE GUTIÉRREZ-RODRÍGUEZ1, ELENA G. GONZALEZ1, ÍÑIGO 6 MARTÍNEZ-SOLANO2,3,* 7 1 Museo Nacional de Ciencias Naturales, CSIC, c/ José Gutiérrez Abascal, 2, 28006 8 Madrid, Spain; 2 Instituto de Investigación en Recursos Cinegéticos, CSIC-UCLM- 9 JCCM, Ronda de Toledo, s/n, 13005 Ciudad Real, Spain; 3 (present address: Centro de 10 Investigação em Biodiversidade e Recursos Genéticos (CIBIO), Rua Padre Armando 11 Quintas, s/n, 4485-661 Vairão, Portugal 12 Number of words: 3017; abstract: 138 13 (*) Corresponding author: 14 I. Martínez-Solano 15 Instituto de Investigación en Recursos Cinegéticos (CSIC-UCLM-JCCM) 16 Ronda de Toledo, s/n 17 13005 Ciudad Real, Spain 18 Phone: +34 926 295 450 ext. 6255 19 Fax: +34 926 295 451 20 Email: [email protected] 21 22 Running title: Microsatellite loci in Pleurodeles waltl 23 1 24 Abstract 25 Twelve novel polymorphic microsatellite loci were isolated and characterized for the 26 Iberian Ribbed Newt, Pleurodeles waltl (Caudata, Salamandridae). The distribution of 27 this newt ranges from central and southern Iberia to northwestern Morocco. 28 Polymorphism of these novel loci was tested in 40 individuals from two Iberian 29 populations and compared with previously published markers. The number of alleles 30 per locus ranged from two to eight. Observed and expected heterozygosity ranged from 31 0.13 to 0.57 and from 0.21 to 0.64, respectively. -

Zootaxa, Caudata, Pleurodeles
Zootaxa 488: 1–24 (2004) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ ZOOTAXA 488 Copyright © 2004 Magnolia Press ISSN 1175-5334 (online edition) Taxonomic revision of Algero-Tunisian Pleurodeles (Caudata: Salamandridae) using molecular and morphological data. Revalidation of the taxon Pleurodeles nebulosus (Guichenot, 1850) SALVADOR CARRANZA1* & EDWARD WADE2 1. Department of Zoology, The Natural History Museum, London, SW7 5BD ([email protected]) * Present address: Departament de Biologia Animal, Universitat de Barcelona, Av. Diagonal 645, E-08028 Barcelona, Spain ([email protected]) 2. Middlesex University, Cat Hill, Barnet, Hertfordshire, EN4 8HT ([email protected]) Abstract The taxonomic status of Algero-Tunisian Pleurodeles was reanalysed in the light of new molecular and morphological evidence. Mitochondrial DNA sequences (396 bp of the cytochrome b and 369 of the 12S rRNA) and the results of the morphometric analysis, indicate that Algero-Tunisian P. poireti consists of two genetically and morphologically distinct forms. One restricted to the Edough Peninsula, and another one covering all the rest of its distribution in Algeria and Tunisia. The name P. p oire ti (Gervais, 1835) is restricted to the population of the Edough Peninsula, while P. nebulous (Guichenot, 1850) correctly applies to all other populations in the distribution. P. po ire ti originated approximately 4.2 Myr ago, probably as a result of the Edough Peninsula being a Pliocene fossil island, allowing both forms of Algero-Tunisian Pleurodeles to diverge both genetically and mor- phologically. Key words: Pleurodeles, Algeria, taxonomy, mitochondrial DNA, 12S rRNA, cytochrome b, mor- phology, Pliocene fossil island Introduction The genus Pleurodeles currently consists of two species. -

Amphibians and Reptiles of the Mediterranean Basin
Chapter 9 Amphibians and Reptiles of the Mediterranean Basin Kerim Çiçek and Oğzukan Cumhuriyet Kerim Çiçek and Oğzukan Cumhuriyet Additional information is available at the end of the chapter Additional information is available at the end of the chapter http://dx.doi.org/10.5772/intechopen.70357 Abstract The Mediterranean basin is one of the most geologically, biologically, and culturally complex region and the only case of a large sea surrounded by three continents. The chapter is focused on a diversity of Mediterranean amphibians and reptiles, discussing major threats to the species and its conservation status. There are 117 amphibians, of which 80 (68%) are endemic and 398 reptiles, of which 216 (54%) are endemic distributed throughout the Basin. While the species diversity increases in the north and west for amphibians, the reptile diversity increases from north to south and from west to east direction. Amphibians are almost twice as threatened (29%) as reptiles (14%). Habitat loss and degradation, pollution, invasive/alien species, unsustainable use, and persecution are major threats to the species. The important conservation actions should be directed to sustainable management measures and legal protection of endangered species and their habitats, all for the future of Mediterranean biodiversity. Keywords: amphibians, conservation, Mediterranean basin, reptiles, threatened species 1. Introduction The Mediterranean basin is one of the most geologically, biologically, and culturally complex region and the only case of a large sea surrounded by Europe, Asia and Africa. The Basin was shaped by the collision of the northward-moving African-Arabian continental plate with the Eurasian continental plate which occurred on a wide range of scales and time in the course of the past 250 mya [1]. -
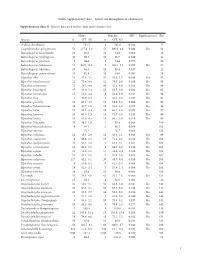
I Online Supplementary Data – Sexual Size Dimorphism in Salamanders
Online Supplementary data – Sexual size dimorphism in salamanders Supplementary data S1. Species data used in this study and references list. Males Females SSD Significant test Ref Species n SVL±SD n SVL±SD Andrias davidianus 2 532.5 8 383.0 -0.280 12 Cryptobranchus alleganiensis 53 277.4±5.2 52 300.9±3.4 0.084 Yes 61 Batrachuperus karlschmidti 10 80.0 10 84.8 0.060 26 Batrachuperus londongensis 20 98.6 10 96.7 -0.019 12 Batrachuperus pinchonii 5 69.6 5 74.6 0.070 26 Batrachuperus taibaiensis 11 92.9±12.1 9 102.1±7.1 0.099 Yes 27 Batrachuperus tibetanus 10 94.5 10 92.8 -0.017 12 Batrachuperus yenyuadensis 10 82.8 10 74.8 -0.096 26 Hynobius abei 24 57.8±2.1 34 55.0±1.2 -0.048 Yes 92 Hynobius amakusaensis 22 75.4±4.8 12 76.5±3.6 0.014 No 93 Hynobius arisanensis 72 54.3±4.8 40 55.2±4.8 0.016 No 94 Hynobius boulengeri 37 83.0±5.4 15 91.5±3.8 0.102 Yes 95 Hynobius formosanus 15 53.0±4.4 8 52.4±3.9 -0.011 No 94 Hynobius fuca 4 50.9±2.8 3 52.8±2.0 0.037 No 94 Hynobius glacialis 12 63.1±4.7 11 58.9±5.2 -0.066 No 94 Hynobius hidamontanus 39 47.7±1.0 15 51.3±1.2 0.075 Yes 96 Hynobius katoi 12 58.4±3.3 10 62.7±1.6 0.073 Yes 97 Hynobius kimurae 20 63.0±1.5 15 72.7±2.0 0.153 Yes 98 Hynobius leechii 70 61.6±4.5 18 66.5±5.9 0.079 Yes 99 Hynobius lichenatus 37 58.5±1.9 2 53.8 -0.080 100 Hynobius maoershanensis 4 86.1 2 80.1 -0.069 101 Hynobius naevius 72.1 76.7 0.063 102 Hynobius nebulosus 14 48.3±2.9 12 50.4±2.1 0.043 Yes 96 Hynobius osumiensis 9 68.4±3.1 15 70.2±3.0 0.026 No 103 Hynobius quelpaertensis 41 52.5±3.8 4 61.3±4.1 0.167 Yes 104 Hynobius -
![Guichenot, 1850] (Amphibia: Salamandridae) in Algeria, with a New Elevational Record for the Species](https://docslib.b-cdn.net/cover/9631/guichenot-1850-amphibia-salamandridae-in-algeria-with-a-new-elevational-record-for-the-species-1559631.webp)
Guichenot, 1850] (Amphibia: Salamandridae) in Algeria, with a New Elevational Record for the Species
Herpetology Notes, volume 14: 927-931 (2021) (published online on 24 June 2021) A new provincial record and an updated distribution map for Pleurodeles nebulosus [Guichenot, 1850] (Amphibia: Salamandridae) in Algeria, with a new elevational record for the species Idriss Bouam1,* and Salim Merzougui2 Pleurodeles Michahelles, 1830, commonly known (1885) noted its presence, very probably mistakenly, as ribbed newts, is an endemic genus of the Ibero- from Biskra, which is an arid region located south of the Maghrebian region, with three species described: P. Saharan Atlas and is abiotically unsuitable for this newt nebulosus (Guichenot, 1850), P. poireti (Gervais, species (see Ben Hassine and Escoriza, 2017; Achour 1835), and P. waltl Michahelles, 1830 (Frost, 2021). and Kalboussi, 2020). We here report the presence of a Pleurodeles nebulosus is an Algero-Tunisian endemic seemingly well-established population of P. nebulosus restricted to a very narrow latitudinal range. It is found in the province of Bordj Bou Arreridj and provide (i) throughout the humid, sub-humid and, to a lesser the first record of the species for this province, thereby extent, semi-arid areas of the northern parts of the extending its known geographic distributional range; two countries, excluding the Edough Peninsula and its (ii) the highest-ever reported elevational record for the surrounding lowlands in northeastern Algeria, where species; and (iii) an updated distribution map of this it is replaced by its sister species P. poireti (Carranza species in Algeria. and Wade, 2004; Escoriza and Ben Hassine, 2019). On 28 April 2020, at 15:30 h, S.M. encountered an Until the end of the 20th century, the known localities individual Pleurodeles nebulosus (Fig. -

Diet of Larval Pleurodeles Waltl (Urodela: Salamandridae) Throughout Its Distributional Range
Limnetica, 39(2): 667-676 (2020). DOI: 10.23818/limn.39.43 © Asociación Ibérica de Limnología, Madrid. Spain. ISSN: 0213-8409 Diet of larval Pleurodeles waltl (Urodela: Salamandridae) throughout its distributional range Daniel Escoriza1,2,*, Jihène Ben Hassine3, Dani Boix2 and Jordi Sala2 1 Institut Català de la Salut. Gran Via de les Corts Catalanes, 587−589, 08004 Barcelona, Spain. 2 GRECO, Institute of Aquatic Ecology, University of Girona, Campus Montilivi, 17071 Girona, Spain. 3 Faculty of Sciences of Tunis, Department of Biology, University of Tunis-El Manar, 2092 Tunis, Tunisia. * Corresponding author: [email protected] Received: 26/11/18 Accepted: 08/10/19 ABSTRACT Diet of larval Pleurodeles waltl (Urodela: Salamandridae) throughout its distributional range Larval diet has important implications for assessing suitable reproduction habitats for amphibians. In this study we investigated the diet of larvae of the Iberian ribbed newt (Pleurodeles waltl), an urodele endemic of the western Mediterranean region. We examined the stomach contents of 150 larvae captured in 30 ponds in Spain, Portugal and Morocco. We found that the larvae predate primarily on microcrustaceans (Cladocera, Ostracoda and Copepoda) and larvae of aquatic insects (Chironomidae, Culicidae and Dytiscidae). However, P. waltl was found to have a broad dietary range, including terrestrial Arthropoda (Homoptera, Sminthuridae and Formicidae), Gastropoda (Physidae and Planorbidae) and amphibian larvae (Anura and Urode- la). As expected, larger larvae had a more diverse diet, as they can capture larger prey. Key words: insect larvae, microcrustaceans, Morocco, ribbed newt, trophic range RESUMEN Dieta larvaria de Pleurodeles waltl (Urodela: Salamandridae) a través de su rango de distribución La dieta larvaria tiene implicaciones importantes para evaluar la idoneidad de los hábitats de reproducción de los anfibios. -

Pleurodeles Waltl Genome Reveals Novel Features of Tetrapod Regeneration
Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Elewa, A., H. Wang, C. Talavera-López, A. Joven, G. Brito, A. Kumar, L. S. Hameed, et al. 2017. “Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration.” Nature Communications 8 (1): 2286. doi:10.1038/ s41467-017-01964-9. http://dx.doi.org/10.1038/s41467-017-01964-9. Published Version doi:10.1038/s41467-017-01964-9 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:34652068 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ARTICLE DOI: 10.1038/s41467-017-01964-9 OPEN Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration Ahmed Elewa1, Heng Wang2, Carlos Talavera-López1,7, Alberto Joven1, Gonçalo Brito 1, Anoop Kumar1, L. Shahul Hameed1, May Penrad-Mobayed3, Zeyu Yao1, Neda Zamani4, Yamen Abbas5, Ilgar Abdullayev1,6, Rickard Sandberg 1,6, Manfred Grabherr4, Björn Andersson 1 & András Simon1 Salamanders exhibit an extraordinary ability among vertebrates to regenerate complex body 1234567890 parts. However, scarce genomic resources have limited our understanding of regeneration in adult salamanders. Here, we present the ~20 Gb genome and transcriptome of the Iberian ribbed newt Pleurodeles waltl, a tractable species suitable for laboratory research. -

Chapter 27 Conservation Status of Amphibians in Tunisia
Basic and Applied Herpetology 27 (2013): 85-100 Chapter 27 Conservation status of amphibians in Tunisia Nabil Amor 1,2,* , Mohsen Kalboussi 3, Khaled Said 2 1 Center for Functional and Evolutionary Ecology and Biogeography of Vertebrates (EPHE), UMR5175 – CNRS Montpellier, France. 2 Unité de Recherche: Génétique, Biodiversité et Valorisation des Bioresources, Institut Supérieur de Biotechnologie de Monastir, Monastir, Tunisia. 3 Institut Sylvo-Pastoral, Tabarka, Tunisia. Authors are listed in alphabetical order. * Correspondence: Unité de Recherche: Génétique, Biodiversité et Valorisation des Bioresources, UR/09-30, Institut Supérieur de Biotechnologie de Monastir, Monastir 5000, Tunisia. Phone: +216 99904622, Fax: +216 71573526. Email: [email protected] Received: 10 January 2013; received in revised form: 17 September 2013; accepted: 18 September 2013. The North African amphibian fauna was once regarded as limited in diversity, but increased field and laboratory research in the region has subsequently revealed considerable endemism and data such as these are necessary for making objec - tive and justifiable recommendations for conservation. Our research, coupled with findings from the literature, allow an up-to-date analysis of distribution, status of populations, and actual and potential threats to the continued survival of all species within Tunisia. The Tunisian batrachofauna currently consists of seven species grouped in seven genera: Pleurodeles , Bufotes , Discoglossus , Bufo , Amietophrynus , Pelophylax , and Hyla . Whereas other species are characterized by wider distributions from north to south, Bufo spinosus appears restricted to the mountainous northwestern corner where major protected areas occur. Pleurodeles nebulosus and Hyla meridionalis appear restricted to humid, subhumid, and semi-arid localities in northern Tunisia, in the Khroumirie region, but also within the Mogod region, around Tunis and the Cap Bon Peninsula. -

Diversity of Guilds of Amphibian Larvae in North-Western Africa
RESEARCH ARTICLE Diversity of Guilds of Amphibian Larvae in North-Western Africa Daniel Escoriza1*, Jihène Ben Hassine2,3 1 GRECO, Institute of Aquatic Ecology, University of Girona, Campus Montilivi, Girona, Spain, 2 Faculty of Sciences of Tunis, University Tunis El Manar, Tunis, Tunisia, 3 Laboratory Ecology, Biodiversity and Environment, Faculty of Sciences of TeÂtouan, University Abdelmalek EssaaÃdi, El M'Hannech II, Tetouan, Morocco * [email protected] Abstract a1111111111 The composition and diversity of biotic assemblages is regulated by a complex interplay of a1111111111 environmental features. We investigated the influence of climate and the aquatic habitat a1111111111 a1111111111 conditions on the larval traits and the structure of amphibian larval guilds in north-western a1111111111 Africa. We classified the species into morphological groups, based on external traits: body shape, size, and the relative positions of the eyes and oral apparatus. We characterized the guild diversity based on species richness and interspecific phylogenetic/functional relation- ships. The larvae of the urodeles were classified as typical of either the stream or pond type, OPEN ACCESS and the anurans as typical of either the lentic-benthic or lentic-nektonic type. The variations in the body shapes of both urodeles and anurans were associated with the type of aquatic habitat Citation: Escoriza D, Ben Hassine J (2017) Diversity of Guilds of Amphibian Larvae in North- (lentic vs lotic) and the types of predators present. Most of the urodele guilds (98.9%) con- Western Africa. PLoS ONE 12(1): e0170763. tained a single species, whereas the anuran guilds were usually more diverse. Both the phylo- doi:10.1371/journal.pone.0170763 genetic and functional diversity of the anuran guilds were positively influenced by the size of Editor: Benedikt R. -
Reciprocal Role of Salamanders in Aquatic Energy Flow Pathways
diversity Review Reciprocal Role of Salamanders in Aquatic Energy Flow Pathways Javier Sánchez-Hernández Área de Biodiversidad y Conservación, Departamento de Biología y Geología, Física y Química Inorgánica, Universidad Rey Juan Carlos, Móstoles, 28933 Madrid, Spain; [email protected]; Tel.: +34-914-888-158 Received: 19 November 2019; Accepted: 15 January 2020; Published: 17 January 2020 Abstract: Many species of salamanders (newts and salamanders per se) have a pivotal role in energy flow pathways as they include individuals functioning as prey, competitors, and predators. Here, I synthesize historic and contemporary research on the reciprocal ecological role of salamanders as predators and prey in aquatic systems. Salamanders are a keystone in ecosystem functioning through a combination of top–down control, energy transfer, nutrient cycling processes, and carbon retention. The aquatic developmental stages of salamanders are able to feed on a wide variety of invertebrate prey captured close to the bottom as well as on small conspecifics (cannibalism) or other sympatric species, but can also consume terrestrial invertebrates on the water surface. This capacity to consume allochthonous resources (terrestrial invertebrates) highlights the key role of salamanders as couplers of terrestrial and aquatic ecosystems (i.e., aquatic–terrestrial linkages). Salamanders are also an important food resource for other vertebrates such as fish, snakes, and mammals, covering the energy demands of these species at higher trophic levels. This study emphasizes the ecological significance of salamanders in aquatic systems as central players in energy flow pathways, enabling energy mobility among trophic levels (i.e., vertical energy flow) and between freshwater and terrestrial habitats (i.e., lateral energy flow). -
Amphibians of Algeria: New Data on the Occurrence and Natural History
RESEARCH ARTICLE The Herpetological Bulletin 142, 2017: 6-18 Amphibians of Algeria: New data on the occurrence and natural history JIHÈNE BEN HASSINE1,2* & DANIEL ESCORIZA2,3 1Faculty of Sciences of Tunis, Department of Biology, University of Tunis-El Manar. 2092 Tunis, Tunisia 2Laboratory Ecology, Biodiversity and Environment, University Abdelmalek Essaâdi, El M’Hannech II, 93030, Tétouan, Morocco 3Institut Català de la Salut. Gran Via de les Corts Catalanes, 587−589, 08004 Barcelona, Spain *Corresponding author Email: [email protected] ABSTRACT - Algeria is a country from the Maghreb with a little known batrachofauna. In order to improve knowledge of the distribution of amphibians in this country we carried out several surveys in northern Algeria between 2010 and 2017. Maps with original data on the distribution ranges and niche model for every species have been made for the first time. This includes original data on breeding phenology, breeding habitat and terrestrial habitat features of the observed species. Our data indicated that several species could be more widespread in Algeria than previously suggested. The apparent discontinuity of their ranges and the supposed rarity of some species such as Pleurodeles nebulosus, Salamandra algira algira and Bufo spinosus is likely due to a lack of previous survey effort. Some species, such as Discoglossus pictus, Pelophylax saharicus, Hyla meridionalis and Sclerophrys mauritanica are widely distributed and abundant in the studied region. Our results confirmed the presence of several species in historical sites, but also the presence of numerous new populations. Some historical records of P. nebulosus, S. algira algira and H. meridionalis were not confirmed.