Short Note Development and Characterization of Twelve New
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

PART 2/2. Encl.: SWD(2021) 204 Final
Council of the European Union Brussels, 19 July 2021 (OR. en) 10943/21 ADD 1 VETER 66 ENV 540 RECH 361 COVER NOTE From: Secretary-General of the European Commission, signed by Ms Martine DEPREZ, Director date of receipt: 14 July 2021 To: Mr Jeppe TRANHOLM-MIKKELSEN, Secretary-General of the Council of the European Union No. Cion doc.: SWD(2021) 204 final - PART 2/2 Subject: COMMISSION STAFF WORKING DOCUMENT Summary Report on the statistics on the use of animals for scientific purposes in the Member States of the European Union and Norway in 2018 Delegations will find attached document SWD(2021) 204 final - PART 2/2. Encl.: SWD(2021) 204 final - PART 2/2 10943/21 ADD 1 OT/pj LIFE.3 EN EUROPEAN COMMISSION Brussels, 14.7.2021 SWD(2021) 204 final PART 2/2 COMMISSION STAFF WORKING DOCUMENT Summary Report on the statistics on the use of animals for scientific purposes in the Member States of the European Union and Norway in 2018 EN EN PART C: MEMBER STATE DATA 2018 MEMBER STATE COMPARATIVE TABLES FOR 2018 MEMBER STATE DATA 2018 .............................................................................................. 10 VI Member State narratives and data submissions 2018 ......................................................... 10 VI.1. Introduction..................................................................................................................... 10 VI.2. Member State narratives and data submissions for 2018 ............................................... 11 Austria ..................................................................................................................................... -

Cop18 Prop. 39
Original language: English CoP18 Prop. 39 CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA ____________________ Eighteenth meeting of the Conference of the Parties Colombo (Sri Lanka), 23 May – 3 June 2019 CONSIDERATION OF PROPOSALS FOR AMENDMENT OF APPENDICES I AND II A. Proposal Inclusion of Echinotriton chinhaiensis (Chang, 1932) and Echinotriton maxiquadratus Hou, Wu, Yang, Zheng, Yuan, and Li, 2014, both of which are endemic to China in Appendix Ⅱ, in accordance with Article Ⅱ, paragraph 2 (a) of the Convention and satisfying Criterion B in Annex 2a of Resolution Conf. 9.24 (Rev. CoP17). The international trade of these two newts should be monitored to minimise the impact of illegal hunting driven by international pet trade or collection on the survival of these two critically endangered species B. Proponent China*: C. Supporting statement 1. Taxonomy 1.1 Class: Amphibia 1.2 Order: Caudata 1.3 Family: Salamandridae 1.4 Genus, species or subspecies, including author and year: 1.5 Scientific synonyms: Echinotriton chinhaiensis: Tylototriton chinhaiensis Chang, 1932; Tylototriton (Echinotriton) chinhaiensis; Pleurodeles chinhaiensis (Chang, 1932); Pleurodeles (Tylototrion) chinhaiensis 1.6 Common names: English: E. chinhaiensis: Chinhai Spiny Newt, Chinhai Spiny Crocodile Newt E. maxiquadratus: Mountain Spiny Newt, Mountain Spiny Crocodile Newt French: Spanish: 1.7 Code numbers: N/A * The geographical designations employed in this document do not imply the expression of any opinion whatsoever on the part of the CITES Secretariat (or the United Nations Environment Programme) concerning the legal status of any country, territory, or area, or concerning the delimitation of its frontiers or boundaries. The responsibility for the contents of the document rests exclusively with its author. -

Reproductive Biology and Endocrinology Biomed Central
Reproductive Biology and Endocrinology BioMed Central Research Open Access Lifelong testicular differentiation in Pleurodeles waltl (Amphibia, Caudata) Stéphane Flament*, Hélène Dumond, Dominique Chardard and Amand Chesnel Address: EA 3442 Aspects cellulaires et moléculaires de la reproduction et du développement, Nancy-Université, Faculté des Sciences, Boulevard des Aiguillettes, BP 70239, 54506 Vandoeuvre-les-Nancy, France Email: Stéphane Flament* - [email protected]; Hélène Dumond - [email protected]; Dominique Chardard - [email protected]; Amand Chesnel - [email protected] * Corresponding author Published: 5 March 2009 Received: 10 December 2008 Accepted: 5 March 2009 Reproductive Biology and Endocrinology 2009, 7:21 doi:10.1186/1477-7827-7-21 This article is available from: http://www.rbej.com/content/7/1/21 © 2009 Flament et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: In numerous Caudata, the testis is known to differentiate new lobes at adulthood, leading to a multiple testis. The Iberian ribbed newt Pleurodeles waltl has been studied extensively as a model for sex determination and differentiation. However, the evolution of its testis after metamorphosis is poorly documented. Methods: Testes were obtained from Pleurodeles waltl of different ages reared in our laboratory. Testis evolution was studied by several approaches: morphology, histology, immunohistochemistry and RT-PCR. Surgery was also employed to study testis regeneration. -

Cytogenetic and Genetic Evidence of Male Sexual Inversion by Heat Treatment in the Newt Pleurodeles Poireti
CHROMOSOMA Chromosoma (Bet1) (1984) 90:261-264 Springer-Verlag 1984 Cytogenetic and genetic evidence of male sexual inversion by heat treatment in the newt Pleurodeles poireti C. Dournon 1, F. Guillet 1, D. Boucher 2, and J.C. Lacroix 2 1 Laboratoire de Biologic Animale; 2Laboratoire de G6n~tique du D~veloppement, Universit~ P. et M. Curie, 9 quai St Bernard 75230 Paris Cedex 05, France Abstract. Larvae of Pleurodeles poireti were maintained the lengthy reproductive cycle of the amphibians. It should during their development at a high temperature (31 ~ C). be interesting therefore to identify the genotypic sex of an In several species of amphibians, such a treatment is known experimental animal by cytogenetic or other genetic criteria, to change the sex ratio through the inversion of genotypic i.e. through sex chromosomes or sex-linked characters re- females into phenotypic males. Pleurodeles poireti is an ex- spectively. ception. It is the first reported amphibian in which heat In a geographical race of P. poireti, the sex chromo- induces an inversion of genotypic males into functional phe- somes can be distinguished in preparations of lampbrush notypic females. The sexual genotype of standard and ex- chromosomes of the oocytes (Lacroix 1970). Moreover, perimental phenotypic females was determined through he- Ferrier et al. (1980, 1983), have shown that in P. waltlii terochromosomes in lampbrush stage. In the present study, the enzyme peptidase I shows a sex-linked polymorphism. we have utilised another technique for identification of sex- The effects of heat treatment on sexual differentiation ual genotype, applicable to both phenotypic males and fe- of larvae of P. -

Salamander Genome Gives Clues About Unique Regenerative Ability 22 December 2017
Salamander genome gives clues about unique regenerative ability 22 December 2017 Molecular Biology. "What's needed now are functional studies of these microRNA molecules to understand their function in regeneration. The link to cancer cells is also very interesting, especially bearing in mind newts' marked resistance to tumour formation." Even though the abundance of stem cell microRNA genes is quite surprising, it alone cannot explain how salamanders regenerate so well. Professor Simon predicts that the explanation lies in a combination of genes unique to salamanders and how other more common genes orchestrate and control the actual regeneration process. One of the reasons why salamander genomes have not been sequenced before is its sheer size - six Credit: CC0 Public Domain times bigger than the human genome in the case of the Iberian newt, which has posed an enormous technical and methodological challenge. Researchers at Karolinska Institutet in Sweden "It's only now that the technology is available to have managed to sequence the giant genome of a handle such a large genome," says Professor salamander, the Iberian ribbed newt, which is a full Simon. "The sequencing per se doesn't take that six times greater than the human genome. long - it's recreating the genome from the Amongst the early findings is a family of genes that sequences that's so time consuming." can provide clues to the unique ability of salamanders to rebuild complex tissue, even body "We all realised how challenging it was going to parts. The study is published in Nature be," recounts first author Ahmed Elewa, Communications. -

50 CFR Ch. I (10–1–20 Edition) § 16.14
§ 15.41 50 CFR Ch. I (10–1–20 Edition) Species Common name Serinus canaria ............................................................. Common Canary. 1 Note: Permits are still required for this species under part 17 of this chapter. (b) Non-captive-bred species. The list 16.14 Importation of live or dead amphib- in this paragraph includes species of ians or their eggs. non-captive-bred exotic birds and coun- 16.15 Importation of live reptiles or their tries for which importation into the eggs. United States is not prohibited by sec- Subpart C—Permits tion 15.11. The species are grouped tax- onomically by order, and may only be 16.22 Injurious wildlife permits. imported from the approved country, except as provided under a permit Subpart D—Additional Exemptions issued pursuant to subpart C of this 16.32 Importation by Federal agencies. part. 16.33 Importation of natural-history speci- [59 FR 62262, Dec. 2, 1994, as amended at 61 mens. FR 2093, Jan. 24, 1996; 82 FR 16540, Apr. 5, AUTHORITY: 18 U.S.C. 42. 2017] SOURCE: 39 FR 1169, Jan. 4, 1974, unless oth- erwise noted. Subpart E—Qualifying Facilities Breeding Exotic Birds in Captivity Subpart A—Introduction § 15.41 Criteria for including facilities as qualifying for imports. [Re- § 16.1 Purpose of regulations. served] The regulations contained in this part implement the Lacey Act (18 § 15.42 List of foreign qualifying breed- U.S.C. 42). ing facilities. [Reserved] § 16.2 Scope of regulations. Subpart F—List of Prohibited Spe- The provisions of this part are in ad- cies Not Listed in the Appen- dition to, and are not in lieu of, other dices to the Convention regulations of this subchapter B which may require a permit or prescribe addi- § 15.51 Criteria for including species tional restrictions or conditions for the and countries in the prohibited list. -

Zootaxa, Caudata, Pleurodeles
Zootaxa 488: 1–24 (2004) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ ZOOTAXA 488 Copyright © 2004 Magnolia Press ISSN 1175-5334 (online edition) Taxonomic revision of Algero-Tunisian Pleurodeles (Caudata: Salamandridae) using molecular and morphological data. Revalidation of the taxon Pleurodeles nebulosus (Guichenot, 1850) SALVADOR CARRANZA1* & EDWARD WADE2 1. Department of Zoology, The Natural History Museum, London, SW7 5BD ([email protected]) * Present address: Departament de Biologia Animal, Universitat de Barcelona, Av. Diagonal 645, E-08028 Barcelona, Spain ([email protected]) 2. Middlesex University, Cat Hill, Barnet, Hertfordshire, EN4 8HT ([email protected]) Abstract The taxonomic status of Algero-Tunisian Pleurodeles was reanalysed in the light of new molecular and morphological evidence. Mitochondrial DNA sequences (396 bp of the cytochrome b and 369 of the 12S rRNA) and the results of the morphometric analysis, indicate that Algero-Tunisian P. poireti consists of two genetically and morphologically distinct forms. One restricted to the Edough Peninsula, and another one covering all the rest of its distribution in Algeria and Tunisia. The name P. p oire ti (Gervais, 1835) is restricted to the population of the Edough Peninsula, while P. nebulous (Guichenot, 1850) correctly applies to all other populations in the distribution. P. po ire ti originated approximately 4.2 Myr ago, probably as a result of the Edough Peninsula being a Pliocene fossil island, allowing both forms of Algero-Tunisian Pleurodeles to diverge both genetically and mor- phologically. Key words: Pleurodeles, Algeria, taxonomy, mitochondrial DNA, 12S rRNA, cytochrome b, mor- phology, Pliocene fossil island Introduction The genus Pleurodeles currently consists of two species. -

Amphibians and Reptiles of the Mediterranean Basin
Chapter 9 Amphibians and Reptiles of the Mediterranean Basin Kerim Çiçek and Oğzukan Cumhuriyet Kerim Çiçek and Oğzukan Cumhuriyet Additional information is available at the end of the chapter Additional information is available at the end of the chapter http://dx.doi.org/10.5772/intechopen.70357 Abstract The Mediterranean basin is one of the most geologically, biologically, and culturally complex region and the only case of a large sea surrounded by three continents. The chapter is focused on a diversity of Mediterranean amphibians and reptiles, discussing major threats to the species and its conservation status. There are 117 amphibians, of which 80 (68%) are endemic and 398 reptiles, of which 216 (54%) are endemic distributed throughout the Basin. While the species diversity increases in the north and west for amphibians, the reptile diversity increases from north to south and from west to east direction. Amphibians are almost twice as threatened (29%) as reptiles (14%). Habitat loss and degradation, pollution, invasive/alien species, unsustainable use, and persecution are major threats to the species. The important conservation actions should be directed to sustainable management measures and legal protection of endangered species and their habitats, all for the future of Mediterranean biodiversity. Keywords: amphibians, conservation, Mediterranean basin, reptiles, threatened species 1. Introduction The Mediterranean basin is one of the most geologically, biologically, and culturally complex region and the only case of a large sea surrounded by Europe, Asia and Africa. The Basin was shaped by the collision of the northward-moving African-Arabian continental plate with the Eurasian continental plate which occurred on a wide range of scales and time in the course of the past 250 mya [1]. -
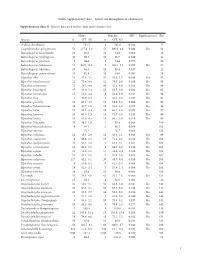
I Online Supplementary Data – Sexual Size Dimorphism in Salamanders
Online Supplementary data – Sexual size dimorphism in salamanders Supplementary data S1. Species data used in this study and references list. Males Females SSD Significant test Ref Species n SVL±SD n SVL±SD Andrias davidianus 2 532.5 8 383.0 -0.280 12 Cryptobranchus alleganiensis 53 277.4±5.2 52 300.9±3.4 0.084 Yes 61 Batrachuperus karlschmidti 10 80.0 10 84.8 0.060 26 Batrachuperus londongensis 20 98.6 10 96.7 -0.019 12 Batrachuperus pinchonii 5 69.6 5 74.6 0.070 26 Batrachuperus taibaiensis 11 92.9±12.1 9 102.1±7.1 0.099 Yes 27 Batrachuperus tibetanus 10 94.5 10 92.8 -0.017 12 Batrachuperus yenyuadensis 10 82.8 10 74.8 -0.096 26 Hynobius abei 24 57.8±2.1 34 55.0±1.2 -0.048 Yes 92 Hynobius amakusaensis 22 75.4±4.8 12 76.5±3.6 0.014 No 93 Hynobius arisanensis 72 54.3±4.8 40 55.2±4.8 0.016 No 94 Hynobius boulengeri 37 83.0±5.4 15 91.5±3.8 0.102 Yes 95 Hynobius formosanus 15 53.0±4.4 8 52.4±3.9 -0.011 No 94 Hynobius fuca 4 50.9±2.8 3 52.8±2.0 0.037 No 94 Hynobius glacialis 12 63.1±4.7 11 58.9±5.2 -0.066 No 94 Hynobius hidamontanus 39 47.7±1.0 15 51.3±1.2 0.075 Yes 96 Hynobius katoi 12 58.4±3.3 10 62.7±1.6 0.073 Yes 97 Hynobius kimurae 20 63.0±1.5 15 72.7±2.0 0.153 Yes 98 Hynobius leechii 70 61.6±4.5 18 66.5±5.9 0.079 Yes 99 Hynobius lichenatus 37 58.5±1.9 2 53.8 -0.080 100 Hynobius maoershanensis 4 86.1 2 80.1 -0.069 101 Hynobius naevius 72.1 76.7 0.063 102 Hynobius nebulosus 14 48.3±2.9 12 50.4±2.1 0.043 Yes 96 Hynobius osumiensis 9 68.4±3.1 15 70.2±3.0 0.026 No 103 Hynobius quelpaertensis 41 52.5±3.8 4 61.3±4.1 0.167 Yes 104 Hynobius -

How Do Adult Songbirds Learn New Sounds? Using Neuromodulators to Probe the Function of the Auditory Association Cortex
University of Massachusetts Amherst ScholarWorks@UMass Amherst Doctoral Dissertations Dissertations and Theses July 2020 How Do Adult Songbirds Learn New Sounds? Using Neuromodulators to Probe the Function of the Auditory Association Cortex Matheus Macedo-Lima University of Massachusetts Amherst Follow this and additional works at: https://scholarworks.umass.edu/dissertations_2 Part of the Behavioral Neurobiology Commons, Comparative and Evolutionary Physiology Commons, Endocrinology Commons, Evolution Commons, Laboratory and Basic Science Research Commons, Molecular and Cellular Neuroscience Commons, and the Systems Neuroscience Commons Recommended Citation Macedo-Lima, Matheus, "How Do Adult Songbirds Learn New Sounds? Using Neuromodulators to Probe the Function of the Auditory Association Cortex" (2020). Doctoral Dissertations. 1947. https://doi.org/10.7275/17542388 https://scholarworks.umass.edu/dissertations_2/1947 This Open Access Dissertation is brought to you for free and open access by the Dissertations and Theses at ScholarWorks@UMass Amherst. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of ScholarWorks@UMass Amherst. For more information, please contact [email protected]. HOW DO ADULT SONGBIRDS LEARN NEW SOUNDS? USING NEUROMODULATORS TO PROBE THE FUNCTION OF THE AUDITORY ASSOCIATION CORTEX A Dissertation Presented by MATHEUS MACEDO-LIMA Submitted to the Graduate School of the University of Massachusetts Amherst in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY May 2020 Neuroscience & Behavior Program © Copyright by Matheus Macedo-Lima 2020 All Rights Reserved HOW DO ADULT SONGBIRDS LEARN NEW SOUNDS? USING NEUROMODULATORS TO PROBE THE FUNCTION OF THE AUDITORY ASSOCIATION CORTEX A Dissertation Presented by MATHEUS MACEDO-LIMA Approved as to style and content by: _________________________________________ Luke Remage-Healey, Chair _________________________________________ Joseph F. -
![Guichenot, 1850] (Amphibia: Salamandridae) in Algeria, with a New Elevational Record for the Species](https://docslib.b-cdn.net/cover/9631/guichenot-1850-amphibia-salamandridae-in-algeria-with-a-new-elevational-record-for-the-species-1559631.webp)
Guichenot, 1850] (Amphibia: Salamandridae) in Algeria, with a New Elevational Record for the Species
Herpetology Notes, volume 14: 927-931 (2021) (published online on 24 June 2021) A new provincial record and an updated distribution map for Pleurodeles nebulosus [Guichenot, 1850] (Amphibia: Salamandridae) in Algeria, with a new elevational record for the species Idriss Bouam1,* and Salim Merzougui2 Pleurodeles Michahelles, 1830, commonly known (1885) noted its presence, very probably mistakenly, as ribbed newts, is an endemic genus of the Ibero- from Biskra, which is an arid region located south of the Maghrebian region, with three species described: P. Saharan Atlas and is abiotically unsuitable for this newt nebulosus (Guichenot, 1850), P. poireti (Gervais, species (see Ben Hassine and Escoriza, 2017; Achour 1835), and P. waltl Michahelles, 1830 (Frost, 2021). and Kalboussi, 2020). We here report the presence of a Pleurodeles nebulosus is an Algero-Tunisian endemic seemingly well-established population of P. nebulosus restricted to a very narrow latitudinal range. It is found in the province of Bordj Bou Arreridj and provide (i) throughout the humid, sub-humid and, to a lesser the first record of the species for this province, thereby extent, semi-arid areas of the northern parts of the extending its known geographic distributional range; two countries, excluding the Edough Peninsula and its (ii) the highest-ever reported elevational record for the surrounding lowlands in northeastern Algeria, where species; and (iii) an updated distribution map of this it is replaced by its sister species P. poireti (Carranza species in Algeria. and Wade, 2004; Escoriza and Ben Hassine, 2019). On 28 April 2020, at 15:30 h, S.M. encountered an Until the end of the 20th century, the known localities individual Pleurodeles nebulosus (Fig. -

Advanced Microinjection Protocol for Gene Manipulation Using the Model
Int. J. Dev. Biol. 63: 281-286 (2019) https://doi.org/10.1387/ijdb.180297th www.intjdevbiol.com Advanced microinjection protocol for gene manipulation using the model newt Pleurodeles waltl TOSHINORI HAYASHI*,1,2,3, MIE NAKAJIMA1, MITSUKI KYAKUNO1,2,3, KANAKO DOI1, IKUMI MANABE1, SHOUHEI AZUMA1 and TAKASHI TAKEUCHI1 1Department of Biomedical Sciences, School of Life Sciences, Faculty of Medicine, Tottori University, Yonago, Tottori, Japan, 2Graduate school of Integrated Science for Life, Hiroshima University, Higashi-Hiroshima, Hiroshima, Japan and 3Amphibian Research Center, Hiroshima University, Higashi-Hiroshima, Hiroshima, Japan. ABSTRACT Urodele amphibian newts have an outstanding history as experimental animals in various research fields such as developmental biology and regeneration biology. We have reported a model experimental system using the Spanish newt, Pleurodeles waltl, and it enables reverse/ molecular genetics through gene manipulation. Microinjection is one of the core techniques in gene manipulation in newts. In the present study, we examined the conditions of the microinjection method, such as egg preparation, de-jelly solution, and formulation of injection medium. We have successfully optimized the injection protocol for P. waltl newts, and our improved protocol is more efficient and lower in cost than previous methods. This protocol can be used for the microinjection of plasmid DNA with I-SceI or mRNA, as well as genome editing using the CRISPR-Cas9 system. This protocol will facilitate research through gene manipulation in newts. KEY WORDS: newt, microinjection, regeneration, transgenesis, genome editing Introduction properties of newts, Gallien and his colleagues found out outstand- ing properties of P. waltl as an experimental animal. After then, Urodele amphibian newts have an outstanding history as an P.