Predicting Future Ocular Chlamydia Trachomatis Infection Prevalence Using Serological, Clinical, Molecular, and Geospatial Data
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-
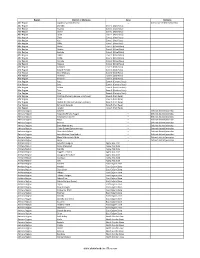
Districts of Ethiopia
Region District or Woredas Zone Remarks Afar Region Argobba Special Woreda -- Independent district/woredas Afar Region Afambo Zone 1 (Awsi Rasu) Afar Region Asayita Zone 1 (Awsi Rasu) Afar Region Chifra Zone 1 (Awsi Rasu) Afar Region Dubti Zone 1 (Awsi Rasu) Afar Region Elidar Zone 1 (Awsi Rasu) Afar Region Kori Zone 1 (Awsi Rasu) Afar Region Mille Zone 1 (Awsi Rasu) Afar Region Abala Zone 2 (Kilbet Rasu) Afar Region Afdera Zone 2 (Kilbet Rasu) Afar Region Berhale Zone 2 (Kilbet Rasu) Afar Region Dallol Zone 2 (Kilbet Rasu) Afar Region Erebti Zone 2 (Kilbet Rasu) Afar Region Koneba Zone 2 (Kilbet Rasu) Afar Region Megale Zone 2 (Kilbet Rasu) Afar Region Amibara Zone 3 (Gabi Rasu) Afar Region Awash Fentale Zone 3 (Gabi Rasu) Afar Region Bure Mudaytu Zone 3 (Gabi Rasu) Afar Region Dulecha Zone 3 (Gabi Rasu) Afar Region Gewane Zone 3 (Gabi Rasu) Afar Region Aura Zone 4 (Fantena Rasu) Afar Region Ewa Zone 4 (Fantena Rasu) Afar Region Gulina Zone 4 (Fantena Rasu) Afar Region Teru Zone 4 (Fantena Rasu) Afar Region Yalo Zone 4 (Fantena Rasu) Afar Region Dalifage (formerly known as Artuma) Zone 5 (Hari Rasu) Afar Region Dewe Zone 5 (Hari Rasu) Afar Region Hadele Ele (formerly known as Fursi) Zone 5 (Hari Rasu) Afar Region Simurobi Gele'alo Zone 5 (Hari Rasu) Afar Region Telalak Zone 5 (Hari Rasu) Amhara Region Achefer -- Defunct district/woredas Amhara Region Angolalla Terana Asagirt -- Defunct district/woredas Amhara Region Artuma Fursina Jile -- Defunct district/woredas Amhara Region Banja -- Defunct district/woredas Amhara Region Belessa -- -

Effects of Fertilization and Harvesting Age on Yield and Quality of Desho
Agriculture, Forestry and Fisheries 2020; 9(4): 113-121 http://www.sciencepublishinggroup.com/j/aff doi: 10.11648/j.aff.20200904.13 ISSN: 2328-563X (Print); ISSN: 2328-5648 (Online) Effects of Fertilization and Harvesting Age on Yield and Quality of Desho (Pennisetum pedicellatum ) Grass Under Irrigation, in Dehana District, Wag Hemra Zone, Ethiopia Awoke Kefyalew 1, Berhanu Alemu 2, Alemu Tsegaye 3 1Department of Animal Science, College of Agriculture, Oda Bultum University, Chiro, Ethiopia 2Department of Animal Science, College of Agriculture and Natural Resources, Debre Markos University, Debre Markos, Ethiopia 3Sekota Dry Land Agricultural Research Center, Sekota, Ethiopia Email address: To cite this article: Awoke Kefyalew, Berhanu Alemu, Alemu Tsegaye. Effects of Fertilization and Harvesting Age on Yield and Quality of Desho (Pennisetum pedicellatum ) Grass Under Irrigation, in Dehana District, Wag Hemra Zone, Ethiopia. Agriculture, Forestry and Fisheries . Vol. 9, No. 4, 2020, pp. 113-121. doi: 10.11648/j.aff.20200904.13 Received : June 5, 2020; Accepted : June 19, 2020; Published : July 28, 2020 Abstract: The experiment was conducted to evaluating the effects of fertilizer and harvesting age on agronomic performance, chemical composition and economic feasibility of Desho (Pennisetum Pedicellatum) grass under irrigation, in Ethiopia. A factorial arrangement with four fertilizer types (control, urea, compost and urea + compost), and three harvesting ages (90, 120 and 150) with three replications were used. Data on morphological characteristics of the grass were recorded. Based on the data collected, harvesting age was significantly affected the agronomic parameters of the grass. Plant height (PH), number of tillers per plant (NTPP), number of leaves per plant (NLPP), number of leaves per tiller (NLPT), dry matter yield (DMY), leaf length (LL) and leaf area (LA) were increased with increasing harvesting age, while leaf to stem ratio (LSR) showed a decreasing trend. -

Ethnobotany, Diverse Food Uses, Claimed Health Benefits And
Shewayrga and Sopade Journal of Ethnobiology and Ethnomedicine 2011, 7:19 http://www.ethnobiomed.com/content/7/1/19 JOURNAL OF ETHNOBIOLOGY AND ETHNOMEDICINE RESEARCH Open Access Ethnobotany, diverse food uses, claimed health benefits and implications on conservation of barley landraces in North Eastern Ethiopia highlands Hailemichael Shewayrga1* and Peter A Sopade2,3 Abstract Background: Barley is the number one food crop in the highland parts of North Eastern Ethiopia produced by subsistence farmers grown as landraces. Information on the ethnobotany, food utilization and maintenance of barley landraces is valuable to design and plan germplasm conservation strategies as well as to improve food utilization of barley. Methods: A study, involving field visits and household interviews, was conducted in three administrative zones. Eleven districts from the three zones, five kebeles in each district and five households from each kebele were visited to gather information on the ethnobotany, the utilization of barley and how barley end-uses influence the maintenance of landrace diversity. Results: According to farmers, barley is the “king of crops” and it is put for diverse uses with more than 20 types of barley dishes and beverages reportedly prepared in the study area. The products are prepared from either boiled/roasted whole grain, raw- and roasted-milled grain, or cracked grain as main, side, ceremonial, and recuperating dishes. The various barley traditional foods have perceived qualities and health benefits by the farmers. Fifteen diverse barley landraces were reported by farmers, and the ethnobotany of the landraces reflects key quantitative and qualitative traits. Some landraces that are preferred for their culinary qualities are being marginalized due to moisture shortage and soil degradation. -

Development Food Assistance Program (DFAP) Fiscal
Food for the Hungry Ethiopia Title II - Development Food Assistance Program (DFAP) Fiscal Year 2016 Annual Results Report Award Number: AID-FFP-A-11-00012 Submission Date: November 21, 2016 Re-Submission Date: March xx, 2017 FH HQ CONTACT FH/ETHIOPIA CONTACT Anthony Koomson Craig Jaggers Global Director, Public Resources Development Country Director Food for the Hungry P.O. Box: 4181 1001 Connecticut Ave. NW Suite 1115 Addis Ababa Washington, DC 20036 Ethiopia Tel: 202-683-7577 Tel: 251-016-188461 Fax: 202-547-0523 Fax: 251-016-188482 Email: [email protected] Email: [email protected] Table of Contents LIST OF ACRONYMS .................................................................................................................IV 1. UPLOADS TO FFPMIS .......................................................................................................... 1 A. ANNUAL RESULTS REPORT (ARR) NARRATIVE 1 I) PROJECT ACTIVITIES AND RESULTS ............................................................................................................................ 1 SO 1: HEALTH AND NUTRITION OF WOMEN AND UNDER FIVE CHILDREN IMPROVED 1 SO 2: COMMUNITY RESILIENCY TO WITHSTAND SHOCKS IMPROVED 9 II) DIRECT PARTICIPANTS BY STRATEGIC OBJECTIVE/PURPOSE .............................................................................. 19 III) CHALLENGES, SUCCESSES, AND LESSONS LEARNED ........................................................................................... 19 B. SUCCESS STORIES 20 C. INDICATOR PERFORMANCE TRACKING TABLE (IPTT) 20 D. INDICATOR -

Evaluation Report
EVALUATION REPORT Program for Emergency Seed support (PESS) project in Wag Himra Zone of Amhara Region Submitted to Food for the Hungry (FH) Karamara Consultancy Service Samuel Lemma February 2021 +251 911 455938; +251 922 785412; +251 984 000804 Addis Ababa Website: http://www.karamaraconsultancy.com Email: [email protected]; [email protected] P.O. Box 4674, Addis Ababa, Contents ACKNOWLEDGMENT........................................................................................................... iv ACRONYMS ............................................................................................................................. v EXECUTIVE SUMMARY ...................................................................................................... vi 1. INTRODUCTION ................................................................................................................. 1 2. PURPOSE AND OBJECTIVES OF THE EVALUATION............................................... 3 2.1 Purpose and general objectives of the Evaluation.......................................................... 3 2.2 Specific Objectives: ....................................................................................................... 3 3. SCOPE OF THE EVALUATION ...................................................................................... 3 3.1 Programme Focus........................................................................................................... 3 3.2 Geographical Focus ....................................................................................................... -

Gender, HIV/AIDS and Food Security Linkage and Integration Into Development Interventions DCG Report No. 32
Gender, HIV/AIDS and Food Security Linkage and Integration into Development Interventions By Dawit Kebede and Solomon Retta December 2004 DCG Report No. 32 Gender, HIV/AIDS and Food Security Linkage and Integration into Development Interventions Dawit Kebede and Solomon Retta DCG Report No. 32 December 2004 The Drylands Coordination Group (DCG) is an NGO-driven forum for exchange of practical experiences and knowledge on food security and natural resource management in the drylands of Africa. DCG facilitates this exchange of experiences between NGOs and research and policy- making institutions. The DCG activities, which are carried out by DCG members in Ethiopia, Eritrea, Mali and Sudan, aim to contribute to improved food security of vulnerable households and sustainable natural resource management in the drylands of Africa. The founding DCG members consist of ADRA Norway, CARE Norway, Norwegian Church Aid, Norwegian People's Aid and The Development Fund. Noragric, the Centre for International Environment and Development Studies at the Agricultural University of Norway, provides the secretariat as a facilitating and implementing body for the DCG. The DCG’s activities are funded by NORAD (the Norwegian Agency for Development Cooperation). Extracts from this publication may only be reproduced after prior consultation with the DCG secretariat. The findings, interpretations and conclusions expressed in this publication are entirely those of the author(s) and cannot be attributed directly to the Drylands Coordination Group. D. Kebede and S. Retta. Drylands Coordination Group Report No. 32 (12, 2004) Drylands Coordination Group c/o Noragric P.O. Box 5003 N-1432 Ås Norway Tel.: +47 64 94 98 23 Fax: +47 64 94 07 60 E-mail: [email protected] Internet: http://www.drylands-group.org ISSN: 1503-0601 Photo credits: T.A. -

Prevalence of Soil-Transmitted Helminths and Schistosoma Mansoni Among a Population-Based Sample of School-Age Children in Amhara Region, Ethiopia Andrew W
Nute et al. Parasites & Vectors (2018) 11:431 https://doi.org/10.1186/s13071-018-3008-0 RESEARCH Open Access Prevalence of soil-transmitted helminths and Schistosoma mansoni among a population-based sample of school-age children in Amhara region, Ethiopia Andrew W. Nute1*, Tekola Endeshaw2, Aisha E. P. Stewart1, Eshetu Sata2, Belay Bayissasse2, Mulat Zerihun2, Demelash Gessesse2, Ambahun Chernet2, Melsew Chanyalew3, Zerihun Tedessse2, Jonathan D. King4, Paul M. Emerson5, E. Kelly Callahan1 and Scott D. Nash1 Abstract Background: From 2011 to 2015, seven trachoma impact surveys in 150 districts across Amhara, Ethiopia, included in their design a nested study to estimate the zonal prevalence of intestinal parasite infections including soil-transmitted helminths (STH) and Schistosoma mansoni. Methods: A multi-stage cluster random sampling approach was used to achieve a population-based sample of children between the ages of 6 and 15 years. Stool samples of approximately 1 g were collected from assenting children, preserved in 10 ml of a sodium acetate-acetic acid-formalin solution, and transported to the Amhara Public Health Research Institute for processing with the ether concentration method and microscopic identification of parasites. Bivariate logistic and negative binomial regression were used to explore associations with parasite prevalence and intensity, respectively. Results: A total of 16,955 children were selected within 768 villages covering 150 districts representing all ten zones of the Amhara region. The final sample included 15,455 children of whom 52% were female and 75% reported regularly attending school. The regional prevalence among children of 6 to 15 years of age was 36.4% (95% confidence interval, CI: 34.9–38.0%) for any STH and 6.9% (95% CI: 5.9–8.1%) for S. -

Gender Responsive Pedagogy: Practices, Challenges & Opportunities
Gender Responsive Pedagogy: Practices, Challenges & Opportunities - A Case of Secondary Schools of North Wollo Zone, Ethiopia Mollaw Abraha1*, Asrat Dagnew2 and Amera Seifu2 1Education and Behavioral Sciences, Woldia University, Post Box No: 400, Ethiopia 2Education and Behavioral Sciences, Bahir Dar University, Post Box No: 1876, Bahir Dar, Ethiopia Abstract The purpose of the current study was to examine the general secondary school (GSS) science teachers’ gender responsive pedagogy (GRP) implementation status. To do so, descriptive survey research design was employed. Teachers, department heads and school principals were taken as respondents of the study comprehensively. And students were also participants of the study conveniently. Questionnaire, interview, and focus group discussion (FGD) were considered as the principal data collection tools. The collected data was analyzed both qualitatively and quantitatively: the data collected through questionnaire was analyzed via mean, std., one sample t-test. And the data gathered by open ended items, interview, and FGD as well was analyzed via words, phrases, statements and narration. The analyzed data noted that GSS science teachers of the North Wollo Zone had facilitated GRP fairly. Thus, they found as effective in relation to language usage, classroom setups, classroom interaction, and addressing sexual harassment. To do so, availability of qualified school supervisors and principals, and realization of new education training policy (NETP) were considered as possible opportunities. However, teachers as well ineffective to prepare gender responsive (GR) lesson plan, to prepare and use GR instructional materials and to have GR management of sexual maturation. This vanity of teachers was sourced from ranges of challenges: economic, culture, school and teacher related. -

Value Chain and Market Assessment
STRENGTHEN PSNP4 INSTITUTIONS AND RESILIENCE VALUE CHAIN AND MARKET ASSESSMENT Strengthen PSNP 4 Institutions and Resilience Value Chain and Market Assessment Original Submission: June21, 2018; Revised: January 15, 2019; Revised on Sep 23/2019 Value Chain And Market Assessment Report for the Strengthen PSNP4 Institutions and Resilience Development Food Security Activity June 2018 Written by Nimona Birhanu and Henry Swira of CARE and Dan Norell of World Vision i Strengthen PSNP 4 Institutions and Resilience Value Chain and Market Assessment Original Submission: June21, 2018; Revised: January 15, 2019; Revised on Sep 23/2019 Acknowledgements The authors would like to thank all farmers, private sector actors, and government staff who took part in the data collection process for value chain commodity selection and anlysis. Finally, this Value Chain and Market Assessment was possible through the generous support of the United States Agency for International Development and the American people. ii Strengthen PSNP 4 Institutions and Resilience Value Chain and Market Assessment Original Submission: June21, 2018; Revised: January 15, 2019; Revised on Sep 23/2019 Table of Contents Acknowledgements .......................................................................................................................................... ii Acronyms .......................................................................................................................................................... vi Executive Summary .................................................................................................................................. -

RFQ for Goods/Services
Section II: Schedule of Requirements / Terms of Reference (ToR) eSourcing reference: ITB/2019/11476 GENERAL DESCRIPTION Service description: Establishment of a Long Term Agreement (LTA) for the Provision of Vehicle Rental Services for all Regional States in Ethiopia except Somali Regional State; Ethiopia Location: All Regional States Zones and Woredas except Somali Regional State - Ethiopia Duration: Long Term Agreement (LTA) to be signed for two (2) years period with a possibility of extension for one more year based on satisfactory performance I. BACKGROUND The United Nations Office for Project Services (UNOPS) is a self-financing organization within the United Nations Family and operates within over sixty (60) countries. It is mandated to serve people in need by expanding the ability of the United Nations, governments and other partners to manage Projects, Infrastructure and Procurement in a sustainable and efficient manner. Within these three core areas of expertise, UNOPS provides its partners with advisory, implementation and transactional services, with projects ranging from building schools and hospitals to procuring goods and services and training local personnel. UNOPS works closely with governments and communities to ensure increased economic, social and environmental sustainability for the projects we support, with a focus on developing national capacity. UNOPS Ethiopia Operational Hub (ETOH) as part of achieving its mission was created covering the three Horn of Africa countries (Ethiopia, Sudan and Djibouti). UNOPS has supported the Government of Ethiopia, UN organizations and other development partners in the country since 2009. Based in Addis Ababa, our team offers a broad range of project management, procurement and infrastructure development, and advisory services, focusing on health, agriculture, social, humanitarian and economic sectors. -

FOOD AID and the POLITICS of NUMBERS in ETHIOPIA (2002-2004) François Enten
tES GAHIERSDU CRASH FOOD AID AND THE POLITICS OF NUMBERSIN ETHIOPIA (zoo 2-2oo4) FrançoisEnten AH_Ethiopie_GB:AH_Ethiopie_GB 11/12/08 11:24 Page1 FOOD AID AND THE POLITICS OF NUMBERS IN ETHIOPIA (2002-2004) François Enten November 2008 - CRASH/Fondation - Médecins Sans Frontières AH_Ethiopie_GB:AH_Ethiopie_GB 11/12/08 11:24 Page2 ALSO IN THE CAHIER DU CRASH IN THE MSF SPEAKING OUT COLLECTION COLLECTION - Salvadoran Refugee Camps in Honduras (1988) Laurence Binet, October 2003-April 2004 - From Ethiopia to Chechnya A collection of articles by François Jean, March 2004 - Genocide of Rwandan Tutsis (1994) Laurence Binet, October 2003-April 2004 - A critique of MSF - France Operation in Darfur (Sudan) - Rwandan refugee camps Zaire and Tanzania Dr. Corinne Danet (MSF), Sophie Delaunay, (1994-1995) Dr. Evelyne Depoortere (Epicentre), Laurence Binet, October 2003-April 2004 Fabrice Weissman (CRASH/Fondation MSF) Billingual French-English in the same publication, - The violence of the new Rwandan regime January 2007 Laurence Binet, October 2003-April 2004 - Humanitarian action in situations of occupation - Hunting and killing of Rwandan Refugee Xavier Crombé Billingual French-English in the same publication, in Zaire-Congo (1996-1997) January 2007 Laurence Binet, October 2003-April 2004 - History of MSF’s interactions with investigations - Famine and forced relocations in Ethiopia and judicial proceedings: (1984-1986) Legal or humanitarian testimony? Laurence Binet, January 2005 Françoise Bouchet Saulnier, Fabien Dubuet Billingual French-English -

ETHIOPIA Food Security Update March 2010
ETHIOPIA Food Security Update March 2010 According to the Joint Government and Humanitarian Figure 1. Current estimated food security conditions, Partners’ Humanitarian Requirement Document, March 2010 released on 2 February 2010, an estimated 5.23 million people will require emergency food assistance through June 2010. This equates to a net food requirement of 290,271 MT, estimated to cost around USD 231.3 million. Onset and distribution of the belg/gu/ganna/sugum rains (February to June) to date has been normal to above normal in most parts of the country that receive these rains. Water availability has improved in several woredas of eastern parts of Amhara, Tigray, Afar, Oromia and Somali regions following the early onset and good distribution of the belg areas although some areas continue to face shortages. Source: FEWS NET and WFP Ethiopia For more information on FEWS NET’s Food Insecurity Severity Scale, please see: www.fews.net/FoodInsecurity Scale Seasonal calendar and critical events timeline Source: FEWS NET FEWS NET Ethiopia FEWS NET Washington FEWS NET is a USAID-funded activity. The authors’ views expressed in this Addis Ababa 1717 H St NW publication do not necessarily reflect the view of the United States Agency Tel: +251 11 662 02 17/18 Washington DC 20006 for International Development or the United States Government. [email protected] [email protected] www.fews.net/ethiopia ETHIOPIA Food Security Update March 2010 Food security overview More than 5 million people continued to experience high to extreme levels of food insecurity as a result of consecutive below‐normal seasonal rains during the past two years.