TGF Beta Signaling Pathways and Microrna Function in the Female Reproductive Tract
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Table 1. Complete Gene Lists and GO Terms from Figure 3C
Supplemental Table 1. Complete gene lists and GO terms from Figure 3C. Path 1 Genes: RP11-34P13.15, RP4-758J18.10, VWA1, CHD5, AZIN2, FOXO6, RP11-403I13.8, ARHGAP30, RGS4, LRRN2, RASSF5, SERTAD4, GJC2, RHOU, REEP1, FOXI3, SH3RF3, COL4A4, ZDHHC23, FGFR3, PPP2R2C, CTD-2031P19.4, RNF182, GRM4, PRR15, DGKI, CHMP4C, CALB1, SPAG1, KLF4, ENG, RET, GDF10, ADAMTS14, SPOCK2, MBL1P, ADAM8, LRP4-AS1, CARNS1, DGAT2, CRYAB, AP000783.1, OPCML, PLEKHG6, GDF3, EMP1, RASSF9, FAM101A, STON2, GREM1, ACTC1, CORO2B, FURIN, WFIKKN1, BAIAP3, TMC5, HS3ST4, ZFHX3, NLRP1, RASD1, CACNG4, EMILIN2, L3MBTL4, KLHL14, HMSD, RP11-849I19.1, SALL3, GADD45B, KANK3, CTC- 526N19.1, ZNF888, MMP9, BMP7, PIK3IP1, MCHR1, SYTL5, CAMK2N1, PINK1, ID3, PTPRU, MANEAL, MCOLN3, LRRC8C, NTNG1, KCNC4, RP11, 430C7.5, C1orf95, ID2-AS1, ID2, GDF7, KCNG3, RGPD8, PSD4, CCDC74B, BMPR2, KAT2B, LINC00693, ZNF654, FILIP1L, SH3TC1, CPEB2, NPFFR2, TRPC3, RP11-752L20.3, FAM198B, TLL1, CDH9, PDZD2, CHSY3, GALNT10, FOXQ1, ATXN1, ID4, COL11A2, CNR1, GTF2IP4, FZD1, PAX5, RP11-35N6.1, UNC5B, NKX1-2, FAM196A, EBF3, PRRG4, LRP4, SYT7, PLBD1, GRASP, ALX1, HIP1R, LPAR6, SLITRK6, C16orf89, RP11-491F9.1, MMP2, B3GNT9, NXPH3, TNRC6C-AS1, LDLRAD4, NOL4, SMAD7, HCN2, PDE4A, KANK2, SAMD1, EXOC3L2, IL11, EMILIN3, KCNB1, DOK5, EEF1A2, A4GALT, ADGRG2, ELF4, ABCD1 Term Count % PValue Genes regulation of pathway-restricted GDF3, SMAD7, GDF7, BMPR2, GDF10, GREM1, BMP7, LDLRAD4, SMAD protein phosphorylation 9 6.34 1.31E-08 ENG pathway-restricted SMAD protein GDF3, SMAD7, GDF7, BMPR2, GDF10, GREM1, BMP7, LDLRAD4, phosphorylation -

The Transcriptional Co-Regulator Jab1 Is Crucial for Chondrocyte
234 Research Article The transcriptional co-regulator Jab1 is crucial for chondrocyte differentiation in vivo Dongxing Chen1, Lindsay A. Bashur1, Bojian Liang1,*, Martina Panattoni2, Keiko Tamai3,`, Ruggero Pardi2 and Guang Zhou1,3,4,§ 1Department of Orthopaedics, Case Western Reserve University, 10900 Euclid Avenue, Cleveland, OH 44106, USA 2San Raffaele University, School of Medicine and Scientific Institute San Raffaele, Milan, Italy 3Department of Genetics, Case Western Reserve University, 10900 Euclid Avenue, Cleveland, OH 44106, USA 4Case Comprehensive Cancer Center, Case Western Reserve University, 10900 Euclid Avenue, Cleveland, OH 44106, USA *Present address: Department of Orthopaedics, China-Japan Union Hospital, Jilin University, Changchun, Jilin Province, People’s Republic of China `Present address: Oncology Research Laboratories, Daiichi Sankyo Co., Ltd.., Tokyo, Japan §Author for correspondence ([email protected]) Accepted 11 November 2012 Journal of Cell Science 126, 234–243 ß 2013. Published by The Company of Biologists Ltd doi: 10.1242/jcs.113795 Summary The evolutionarily conserved transcriptional cofactor Jab1 plays critical roles in cell differentiation, proliferation, and apoptosis by modulating the activity of diverse factors and regulating the output of various signaling pathways. Although Jab1 can interact with the bone morphogenetic protein (BMP) downstream effector Smad5 to repress BMP signaling in vitro, the role of Jab1 in BMP-mediated skeletogenesis in vivo is still poorly understood. As a key regulator of skeletogenesis, BMP signaling regulates the critical Ihh-Pthrp feedback loop to promote chondrocyte hypertrophy. In this study, we utilized the loxP/Cre system to delineate the specific role of Jab1 in cartilage formation. Strikingly, Jab1 chondrocyte-specific knockout Jab1flox/flox; Col2a1-Cre (cKO) mutants exhibited neonatal lethal chondrodysplasia with severe dwarfism. -

The Nuclear Receptor REVERB Represses the Transcription of Growthdifferentiation Factor 10 and 15 Genes in Rat Endometrium Strom
Physiological Reports ISSN 2051-817X ORIGINAL RESEARCH The nuclear receptor REV-ERBa represses the transcription of growth/differentiation factor 10 and 15 genes in rat endometrium stromal cells Lijia Zhao1, Keishiro Isayama1, Huatao Chen1,*, Nobuhiko Yamauchi1, Yasufumi Shigeyoshi2, Seiichi Hashimoto3 & Masa-aki Hattori1 1 Department of Animal and Marine Bioresource Sciences, Graduate School of Agriculture, Kyushu University, Fukuoka, Japan 2 Department of Anatomy and Neurobiology, Kinki University School of Medicine, Osaka, Japan 3 Graduate School of Medicine, The University of Tokyo, Tokyo, Japan Keywords Abstract Circadian clock, decidualization, growth/ differentiation factors, REV-ERBa. Cellular oscillators in the uterus play critical roles in the gestation processes of mammals through entraining of the clock proteins to numerous downstream Correspondence genes, including growth/differentiation factor (Gdf)10 and Gdf15. The expres- Masa-aki Hattori, Department of Animal and sion of Gdf10 and Gdf15 is significantly increased in the uterus during decidu- Marine Bioresource Sciences, Graduate alization, but the mechanism underlying the regulation of Gdf gene expression School of Agriculture, Kyushu University, in the uterus is poorly understood. Here, we focused on the function of the Hakozaki, Higashi-ku, Fukuoka 812-8581, cellular oscillators in the expression of Gdf family by using uterine endome- Japan. Tel: +81-92-642-2938 trial stromal cells (UESCs) isolated from pregnant Per2-dLuc transgenic rats. Fax: +81-92-642-2938 A significant decline of Per2-dLuc bioluminescence activity was induced in E-mail: [email protected] in vitro decidualized UESCs, and concomitantly the expression of canonical clock genes was downregulated. Conversely, the expression of Gdf10 and ⁄ Present address Gdf15 of the Gdf was upregulated. -

Vg1-Nodal Heterodimers Are the Endogenous Inducers of Mesendoderm Tessa G Montague1*, Alexander F Schier1,2,3,4,5*
RESEARCH ARTICLE Vg1-Nodal heterodimers are the endogenous inducers of mesendoderm Tessa G Montague1*, Alexander F Schier1,2,3,4,5* 1Department of Molecular and Cellular Biology, Harvard University, Cambridge, United States; 2Center for Brain Science, Harvard University, Cambridge, United States; 3Broad Institute of MIT and Harvard, Cambridge, United States; 4Harvard Stem Cell Institute, Cambridge, United States; 5FAS Center for Systems Biology, Harvard University, Cambridge, United States Abstract Nodal is considered the key inducer of mesendoderm in vertebrate embryos and embryonic stem cells. Other TGF-beta-related signals, such as Vg1/Dvr1/Gdf3, have also been implicated in this process but their roles have been unclear or controversial. Here we report that zebrafish embryos without maternally provided vg1 fail to form endoderm and head and trunk mesoderm, and closely resemble nodal loss-of-function mutants. Although Nodal is processed and secreted without Vg1, it requires Vg1 for its endogenous activity. Conversely, Vg1 is unprocessed and resides in the endoplasmic reticulum without Nodal, and is only secreted, processed and active in the presence of Nodal. Co-expression of Nodal and Vg1 results in heterodimer formation and mesendoderm induction. Thus, mesendoderm induction relies on the combination of two TGF-beta- related signals: maternal and ubiquitous Vg1, and zygotic and localized Nodal. Modeling reveals that the pool of maternal Vg1 enables rapid signaling at low concentrations of zygotic Nodal. DOI: https://doi.org/10.7554/eLife.28183.001 Introduction *For correspondence: tessa. [email protected] (TGM); The induction of mesoderm and endoderm (mesendoderm) during embryogenesis and embryonic [email protected] (AFS) stem cell differentiation generates the precursors of the heart, liver, gut, pancreas, kidney and other internal organs. -

Context-Dependent Roles in Cell and Tissue Physiology
Downloaded from http://cshperspectives.cshlp.org/ on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press TGF-b and the TGF-b Family: Context-Dependent Roles in Cell and Tissue Physiology Masato Morikawa,1 Rik Derynck,2 and Kohei Miyazono3 1Ludwig Cancer Research, Science for Life Laboratory, Uppsala University, Biomedical Center, SE-751 24 Uppsala, Sweden 2Department of Cell and Tissue Biology, University of California at San Francisco, San Francisco, California 94143 3Department of Molecular Pathology, Graduate School of Medicine, The University of Tokyo, Bunkyo-ku, Tokyo 113-0033, Japan Correspondence: [email protected] The transforming growth factor-b (TGF-b) is the prototype of the TGF-b family of growth and differentiation factors, which is encoded by 33 genes in mammals and comprises homo- and heterodimers. This review introduces the reader to the TGF-b family with its complexity of names and biological activities. It also introduces TGF-b as the best-studied factor among the TGF-b family proteins, with its diversity of roles in the control of cell proliferation and differentiation, wound healing and immune system, and its key roles in pathology, for exam- ple, skeletal diseases, fibrosis, and cancer. lthough initially thought to stimulate cell TGF-b has been well documented in most cell Aproliferation, just like many growth factors, types, and has been best characterized in epithe- it became rapidly accepted that transforming lial cells. The bifunctional and context-depen- growth factor b (TGF-b) is a bifunctional reg- dent nature of TGF-b activities was further con- ulator that either inhibits or stimulates cell pro- firmed in a large variety of cell systems and liferation. -

Activin Receptor Type IIB (RIIB)
Activin Receptor Type IIB (RIIB) Catalog number A0856-10D Supplier United States Biological Members of the TGF-beta superfamily exert their biological effects by binding to heteromeric complexes of a type I and a type II serine-threonine kinase receptor, both of which are essential for signal transduction. Activin receptor type IIB is a type II receptor that mediates multiple signals for transforming growth factor-beta (TGF-beta) family members, including Activin, Nodal, Bmp7, Gdf1, Gdf3, Myostatin (Gdf8), and Gdf11. Through alternative mRNA splicing, multiple ActR-IIB isoforms can also be generated. Applications Suitable for use in ELISA and Western Blot. Other applications not tested. Recommended Dilution . Western Blot: 1ug/ml using recombinant human Activin RIIB Fc Chimera. Optimal dilutions to be determined by the researcher. Storage and Stability Lyophilized powder may be stored at -20°C. Stable for 12 months after receipt at -20°C. Reconstitute with sterile PBS. Aliquot to avoid repeated freezing and thawing. Store at -20°C. For maximum recovery of product, centrifuge the original vial after thawing and prior to removing the cap. Further dilutions can be made in assay buffer. Immunogen Recombinant protein corresponding to Ser19-Thr134 from human Activin Receptor Type IIB, expressed in NSO cells. Swiss/UniProt Accession # Q13705. Formulation Supplied as a lyophilized powder from PBS, pH 7.4, 5% trehalose. Reconstitute with 200ul sterile PBS. Purity Purified by Protein G affinity chromatography from ascites. Specificity Recognizes human Activin Receptor Type IIB. Shows ~5% crossreactivity with recombinant human Activin RIIA. Product Type Mab Source human Isotype IgG1 Grade Affinity Purified Applications E WB Crossreactivity Hu Storage -20°C Powered by TCPDF (www.tcpdf.org). -

Growth/Differentiation Factor 3 Signals Through ALK7 and Regulates Accumulation of Adipose Tissue and Diet-Induced Obesity
Growth/differentiation factor 3 signals through ALK7 and regulates accumulation of adipose tissue and diet-induced obesity Olov Andersson*, Marion Korach-Andre†, Eva Reissmann*, Carlos F. Iba´ n˜ ez*‡, and Philippe Bertolino* *Division of Molecular Neurobiology, Department of Neuroscience, Karolinska Institutet, S-17177 Stockholm, Sweden; and †Receptor Biology Unit, Department of Biosciences and Nutrition, Karolinska Institutet NOVUM, S-14157 Huddinge, Sweden Edited by Robert J. Lefkowitz, Duke University Medical Center, Durham, NC, and approved March 18, 2008 (received for review January 14, 2008) Growth/differentiation factor 3 (GDF3) is highly expressed in tissue and metabolic control has remained unknown. During adipose tissue, and previous overexpression experiments in mice embryogenesis, GDF3 has been shown to signal through a have suggested that it may act as an adipogenic factor under receptor complex comprising the activin type I receptor ALK4, conditions of high lipid load. GDF3 has been shown to signal via the type II receptors ActRIIA or B, and the coreceptor Cripto to activin receptor ALK4 during embryogenesis, but functional recep- control formation of anterior visceral endoderm and mesoderm tors in adipose tissue are unknown. In this study, we show that (11, 12). The receptors that mediate the functions of GDF3 in the .Gdf3؊/؊ mutant mice accumulate less adipose tissue than WT adult have not been characterized animals and show partial resistance to high-fat diet-induced obe- We and others have recently shown that inactivation of the sity despite similar food intake. We also demonstrate that GDF3 Gdf3 gene in mice leads to embryonic lethality in one-third of the can signal via the ALK4-homolog ALK7 and the coreceptor Cripto, mutant embryos because of pregastrulation developmental mal- both of which are expressed in adipose tissue. -
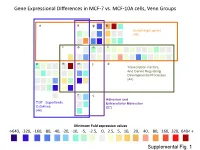
Supplementary Data
Gene Expressional Differences in MCF-7 vs. MCF-10A cells, Venn Groups e k g b Smad target genes (48) l o n i a h m j d Transcription Factors, And Genes Regulating Developmental Processes (44) f c Adhesion and TGF Superfamily Extracellular Molecules Cytokines (27) (44) Minimum Fold expression values ->640, -320, -160, -80, -40, -20, -10, -5, -2.5, 0, 2.5, 5, 10, 20, 40, 80, 160, 320, 640< + Supplemental Fig. 1 Supplemental Table 1. Change of Basal Gene Expressional values in MCF-7 as compared to MCF-10A cell line. Fold T-test Gene # GeneBank Symbol Up/Down p-Value Description /Position Regulation Venn Group a 01 /A01 NM_001105 ACVR1 -4.96 0.0000 Activin A receptor, type I 02 /A02 NM_001616 ACVR2A -2.62 0.0007 Activin A receptor, type IIA 03 /A03 NM_000020 ACVRL1 -1.25 0.7013 Activin A receptor type II-like 1 05 /A05 NM_020547 AMHR2 1.08 0.8043 Anti-Mullerian hormone receptor, type II 16 /B04 NM_004329 BMPR1A -2.40 0.0011 Bone morphogenetic protein receptor, type IA 17 /B05 NM_001203 BMPR1B -7.37 0.0000 Bone morphogenetic protein receptor, type IB 36 /C12 NM_000557 GDF5 -1.38 0.4911 Growth differentiation factor 5 (cartilage-derived morphogenetic protein-1) 37 /D01 NM_001001557 GDF6 1.12 0.9002 Growth differentiation factor 6 38 /D02 NM_182828 GDF7 -1.52 0.4995 Growth differentiation factor 7 53 /E05 NM_020997 LEFTY1 -1.76 0.0529 Left-right determination factor 1 59 /E11 NM_018055 NODAL -3.62 0.1290 Nodal homolog (mouse) 77 /G05 NM_003238 TGFB2 -4.52 0.0566 Transforming growth factor, beta 2 78 /G06 NM_003239 TGFB3 -1.12 0.2902 Transforming -

Genequery™ Human Growth Factors Qpcr Array Kit (GQH-GRF) Catalog #GK123 Product
GeneQuery™ Human Growth Factors qPCR Array Kit (GQH-GRF) Catalog #GK123 Product Description ScienCell's GeneQuery™ Human Growth Factors qPCR Array Kit (GQH-GRF) is designed to facilitate gene expression profiling of 88 human growth factor genes. Growth factors are proteins or hormones which are critical in regulating cellular processes such as apoptosis, proliferation, differentiation, inflammation and wound healing. Brief examples of how genes may be grouped according to their functions are shown below: • Angiopoietins: ANGPT1, ANGPT2 • Bone morphogenetic proteins: BMP1, BMP10, BMP2, BMP4, BMP6, BMP7, BMP8B • Ciliary neurotrophic factor family: CNTF, LIF, IL6 • Colony-stimulating factors: CSF1, CSF2, CSF3 • Epidermal growth factor: EGF • Ephrins: EFNA1, EFNA2, EFNA3, EFNA4, EFNA5, EFNB1, EFNB2, EFNB3 • Fibroblast growth factors: FGF1, FGF10, FGF13, FGF14, FGF16, FGF17, FGF18, FGF19, FGF2, FGF20, FGF23, FGF3, FGF4, FGF5, FGF7, FGF8, FGF9 • GDNF family: GDNF, NRTN, ARTN, PSPN • Growth differentiation factors: GDF1, GDF10, GDF11, GDF2, GDF3, MSTN • Hepatocyte growth factor: HGF • Insulin-like growth factors: IGF1, IGF2 • Interleukins: IL1A, IL1B, IL2, IL3, IL4, IL5, IL6, IL7, IL9, IL10, IL11, IL12B, IL15, IL18 • Neuregulins: NRG1, NRG3 • Neurotrophins: BDNF, NGF, NTF3, NTF4 • Platelet-derived growth factors: PDGFA, PDGFB, PDGFC, PDGFD • Thrombopoietin: THPO • Transforming growth factors: TGFA, TGFB1, TGFB2, TGFB3 • Tumor necrosis factor: TNF • Vascular endothelial growth factors: VEGFA, VEGFB, VEGFC, VEGFD, PGF Note : all gene names follow their official symbols by the Human Genome Organization Gene Nomenclature Committee (HGNC). GeneQuery™ qPCR array kits are qPCR ready in a 96-well plate format, with each well containing one primer set that can specifically recognize and efficiently amplify a target gene's cDNA. -

Insulin Regulates Lipolysis and Fat Mass by Upregulating Growth/Differentiation Factor 3 in Adipose Tissue Macrophages
Diabetes Volume 67, September 2018 1761 Insulin Regulates Lipolysis and Fat Mass by Upregulating Growth/Differentiation Factor 3 in Adipose Tissue Macrophages Yun Bu,1 Katsuhide Okunishi,1 Satomi Yogosawa,1 Kouichi Mizuno,1 Maria Johnson Irudayam,2 Chester W. Brown,2 and Tetsuro Izumi1,3 Diabetes 2018;67:1761–1772 | https://doi.org/10.2337/db17-1201 Previous genetic studies in mice have shown that func- hematopoietic cells inside WAT, and cause chronic inflam- tional loss of activin receptor–like kinase 7 (ALK7), a type I mation and obesity-related disorders (1). The TG content in transforming growth factor-b receptor, increases lipol- adipocytes is determined by the balance between the syn- ysis to resist fat accumulation in adipocytes. Although thesis and breakdown of TG. Although TG synthesis depends growth/differentiation factor 3 (GDF3) has been sug- on the uptake of nutrients, the rate of lipid removal through gested to function as a ligand of ALK7 under nutrient- lipolysis is proportional to the total fat mass as well as the excess conditions, it is unknown how GDF3 production is activities of lipases, and is regulated by external factors, such METABOLISM regulated. Here, we show that a physiologically low level 2 as catecholamine and insulin. It is important to understand of insulin converts CD11c adipose tissue macrophages the mechanisms of fat accumulation to dissect the patho- (ATMs) into GDF3-producing CD11c+ macrophagesexvivo physiology of obesity. Our previous genetic analyses using and directs ALK7-dependent accumulation of fat in vivo. F2 progeny between the Tsumura, Suzuki, obese diabetes Depletion of ATMs by clodronate upregulates adipose (TSOD) and control BALB/c mice revealed a naturally oc- lipases and reduces fat mass in ALK7-intact obese mice, fi curring mutation in Acvr1c encoding the type I transforming but not in their ALK7-de cient counterparts. -

CER1 Is a Common Target of WNT and NODAL Signaling Pathways in Human Embryonic Stem Cells
795-799 24/3/06 13:04 Page 795 INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 17: 795-799, 2006 795 CER1 is a common target of WNT and NODAL signaling pathways in human embryonic stem cells MASUKO KATOH1 and MASARU KATOH2 1M&M Medical BioInformatics, Hongo 113-0033; 2Genetics and Cell Biology Section, National Cancer Center Research Institute, Tokyo 104-0045, Japan Received January 3, 2006; Accepted February 7, 2006 Abstract. Nodal and BMP signaling pathways network with embryogenesis, underwent protein evolution as well as WNT signaling pathway during embryogenesis and carcino- promoter evolution. These facts indicate that molecular genesis. CER1 (Cerberus 1) and GREM3 (CKTSF1B3 or evolution of CER1 orthologs contributes to the significantly CER2) inhibit NODAL signaling through ACVR1B (ALK4) divergent scenarios of early embryogenesis in primates and or ACVR1C (ALK7) to SMAD2 or SMAD3. GREM1 rodents. (CKTSF1B1) inhibits BMP signaling through BMPR1A (ALK3), BMPR1B (ALK6) or ACVR1 (ALK2) to SMAD1, Introduction SMAD5 or SMAD8. CER1, GREM1 and GREM3 are DAN domain (DAND) family members; however, transcriptional TGFB1, TGFB2, TGFB3, NODAL, LEFTY1, LEFTY2, INHA, regulation of DAND family members by canonical WNT INHBA, INHBB, INHBC, INHBE, AMH, BMP2, BMP3, signaling pathway remains unclear. We searched for the BMP4, BMP5, BMP6, BMP7, BMP8A, BMP8B, BMP10, TCF/LEF-binding site within the promoter region of DAND BMP15, GDF1, GDF2, GDF3, GDF5, GDF6, GDF7, GDF8, family genes, including CER1, GREM1, GREM2, GREM3 and GDF9, GDF10, GDF11, and GDF15 are TGFß superfamily NBL1. Because triple TCF/LEF-binding sites were identified genes within the human genome (http://www.gene.ucl.ac.uk). within human CER1 promoter by using bioinformatics and TGFß signals are transduced through type I receptor TGFBR1 human intelligence, comparative genomics analyses on CER1 and type II receptor TGFBR2 to phosphorylate R-SMAD orthologs were further performed. -

Growth/Differentiation Factor 3 Signals Through ALK7 and Regulates Accumulation of Adipose Tissue and Diet-Induced Obesity
Growth/differentiation factor 3 signals through ALK7 and regulates accumulation of adipose tissue and diet-induced obesity Olov Andersson*, Marion Korach-Andre†, Eva Reissmann*, Carlos F. Iba´ n˜ ez*‡, and Philippe Bertolino* *Division of Molecular Neurobiology, Department of Neuroscience, Karolinska Institutet, S-17177 Stockholm, Sweden; and †Receptor Biology Unit, Department of Biosciences and Nutrition, Karolinska Institutet NOVUM, S-14157 Huddinge, Sweden Edited by Robert J. Lefkowitz, Duke University Medical Center, Durham, NC, and approved March 18, 2008 (received for review January 14, 2008) Growth/differentiation factor 3 (GDF3) is highly expressed in tissue and metabolic control has remained unknown. During adipose tissue, and previous overexpression experiments in mice embryogenesis, GDF3 has been shown to signal through a have suggested that it may act as an adipogenic factor under receptor complex comprising the activin type I receptor ALK4, conditions of high lipid load. GDF3 has been shown to signal via the type II receptors ActRIIA or B, and the coreceptor Cripto to activin receptor ALK4 during embryogenesis, but functional recep- control formation of anterior visceral endoderm and mesoderm tors in adipose tissue are unknown. In this study, we show that (11, 12). The receptors that mediate the functions of GDF3 in the .Gdf3؊/؊ mutant mice accumulate less adipose tissue than WT adult have not been characterized animals and show partial resistance to high-fat diet-induced obe- We and others have recently shown that inactivation of the sity despite similar food intake. We also demonstrate that GDF3 Gdf3 gene in mice leads to embryonic lethality in one-third of the can signal via the ALK4-homolog ALK7 and the coreceptor Cripto, mutant embryos because of pregastrulation developmental mal- both of which are expressed in adipose tissue.