Ribosomally Synthesized and Post-Translationally Modified Peptides As Potential Scaffolds for Peptide Engineering
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Dysfunction of the Hypothalamic-Pituitary-Adrenal Axis in HIV Infection and Disease
HORMONES 2008, 7(3):205-216 Review Dysfunction of the Hypothalamic-Pituitary-Adrenal axis in HIV infection and disease Evangelia Zapanti1, Konstantinos Terzidis1, George Chrousos2 11st Department of Endocrinology, “Alexandra” Hospital, Athens, 2Endocrine Unit, Department of Pediatrics, Athens University School of Medicine, “Aghia Sophia” Children’s Hospital, Athens, Greece ABSTRACT Abnormalities of the Hypothalamic-Pituitary-Adrenal (HPA) axis have been documented in HIV patients in the early as well as late stages of the infection and range from subtle subclini- cal disturbances to frank adrenal insufficiency. Potential etiologies of these disorders include opportunistic infections, neoplasms, drugs administered to treat infections, cytokine abnor- malities associated with the HIV disease process and acquired alterations in tissue sensitivity to glucocorticoids. In this article, we present a concise review of HPA abnormalities in HIV infection and disease with regard to their etiology with emphasis on syndromes of hypersen- sitivity/resistance to glucocorticoids associated with antiviral medications and/or the HIV infection itself. Key words: AIDS, Glucocorticoid hypersensitivity, Glucocorticoid resistance, HIV, HPA axis, Vpr INTRODUCTION peptide that plays a central role in coordinating the HPA axis and the systemic response to stress,3 act- HPA axis ing as the main physiologic ACTH stimulus.4 ACTH The HPA axis and the systemic sympathetic/ad- leads to secretion of cortisol (F) and other adrenal renomedullary (sympathoadrenal) system are the steroids, such as dehydroepiandrosterone (DHEA) peripheral branches of the stress system, whose main and aldosterone.5 function is to maintain basal and stress-related ho- meostasis.1 Biologic, physical or psychologic stimuli The HPA axis and the immune-inflammatory activate the stress system, including the HPA axis. -

Endocrinology and Reproduction
Endocrinology and Reproduction Elisabet Stener-Victorin, Professor, PhD Reproductive Endocrinology and Metabolism (REM) group Department of Physiology and Pharmacology Karolinska Institutet, Stockholm, Sweden [email protected] General Consepts of Endocrine Control Hormone – Greek hormaein = ”excite” . Autocrine signalling e.g. interleukin-1 in lymphocytes . Paracrine signalling e.g. growth and clotting factors . Endocrine signalling all circulating hormones Classical Endocrine Organs Other ”non-classical” hormone glands e.g. CNS . Kidney . Stomach . Small intestine . Skin . Heart . Lung . Placenta Katch et al Essentials of Exercise Physiol. Figure 12.1 Hormones Controls and Regulates . Reproduction including gamete production, fertilization, nourishment of the embryo and fetus . Growth and development . Regulates ion and water balance . Regulates cellular metabolism and energy balance . Mobilize the immun system by responding to infection, trauma, and emotional stress Homeostasis . Maintance of steady states by coordinated physiological mechanisms . Contributes to homeostasis by controlling availablity of substrates and metabolism . Regulating body fluid and ion balance Homeostasis – like thermostat in the room Body temperature ~ 37ºC Blood glucose 4.4 – 6.1 mM Ca2+ 4.1 – 5.2 mg/dL Phosphate 0.8 – 1.5 mM How is homestasis achived? Endocrine system : 1. Gland 2. Hormone - receptor 3. Target organ - response Katch et al Essentials of Exercise Physiol. Figure 12.2 Principles for Feed-back Negative Positive (rare) Endocrine Endocrine cell cell A A Target Target Endocrine Endocrine cell cell B B Biological effect Biological effect Principles for Feed-back and Biorythm Complex multilevel . Long feedback loop Hypo- thalamus . Active hormone regulates the hypothalamus Releasing . Short feedback loop hormone . Active hormone regulates pituitary Anterior pituitary Target hormone Target Biorytm - Pulsatile release Endocrine . -

The Adrenal Cortex and Life Gavin P
The Adrenal Cortex and Life Gavin P. Vinson To cite this version: Gavin P. Vinson. The Adrenal Cortex and Life. Molecular and Cellular Endocrinology, Elsevier, 2009, 300 (1-2), pp.2. 10.1016/j.mce.2008.09.008. hal-00532077 HAL Id: hal-00532077 https://hal.archives-ouvertes.fr/hal-00532077 Submitted on 4 Nov 2010 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Accepted Manuscript Title: The Adrenal Cortex and Life Author: Gavin P. Vinson PII: S0303-7207(08)00405-X DOI: doi:10.1016/j.mce.2008.09.008 Reference: MCE 6977 To appear in: Molecular and Cellular Endocrinology Received date: 29-7-2008 Revised date: 4-9-2008 Accepted date: 5-9-2008 Please cite this article as: Vinson, G.P., The Adrenal Cortex and Life, Molecular and Cellular Endocrinology (2008), doi:10.1016/j.mce.2008.09.008 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. -
01 Introduction to Endocrine System.Pdf
ENDOCRINE BLOCK PHYSIOLOGY TEAM 431 Done by : Rand AL-haweal & Abdulaziz Al-Hamad Revised by : Nour Al-Khawajah & Mohammed Asiri Endocrinology (Introduction) Learning objectives: Endocrine vs exocrine gland Transport and clearance Chemical messengers Mechanism of action Hormone Receptors, down-regulation and up-regulation Definition Intracellular signaling Chemical structure Second messenger (cAMP, IP3) Paracrine & autocrine Endocrine vs exocrine gland EXOCRINE GLANDS ENDOCRINE GLANDS Ducts + lumen and surface Chemical messengers + bloodstream Their secretions are released through Their secretions are released directly ducts onto an organ's lumen and surface. into the bloodstream rather than through a duct. Endocrine gland :- ductless, classical gland e.g hypothalamus Endocrine tissue :- tissue secreting hormone e.g skin Chemical messengers The activities of cells, tissues and organs are coordinated by chemical messengers Neurotransmitters Endocrine hormones Neuroendocrine hormones Paracrines :-gland produce affect on local tissue Autocrines Cytokines Juxtacrine :- part of hormone receptor on one cell and other part on other cell. Endocrine glands: Adrenal Pituitary Pancreas Thyroid Ovaries Parathyroid Testes The multiple hormone systems play a key role in regulating almost all body functions: Metabolism Reproduction Growth and development Behavior Water and electrolyte balance Definition: Hormone is a chemical substance released by group of cells to control the function of other type of cells. (It is secreted directly to the blood stream in response to stimulus to cause physiological response at the target tissues.) Types of hormones Affect many different types of cells (eg. GH and Thyroxin) Affect only specific target cells (eg. ACTH and estrogen) What are target cells? Target cells refer to cells that contain specific receptors (binding sites) for a particular hormone. -

ENDOCRINE SYSTEM Hormones •Are Secreted by a Cell Or Group of Cells
ENDOCRINE SYSTEM Hormones •are secreted by a cell or group of cells. •are secreted into the blood •are transported to a distant target. •exert their effect at very low concentrations •act by binding receptors. •action must be terminated. Peptide Hormone Synthesis, Packaging, and Release Messenger RNA on the ribosomes binds amino acids into a peptide chain called a preprohormone. The chain is directed into the ER lumen by a signal sequence of amino acids. Enzymes in the ER chop off the signal sequence, creating an inactiveprohormone. The prohormone passes from the ER through the Golgi complex. Secretory vesicles containing enzymes and prohormone bud off the Golgi. The enzymes chop the prohormone into one or more active peptides plus additional peptide fragments. The secretory vesicle releases its contents by exocytosis into the extracellular space. The hormone moves into the circulation for transport to its target. Steroid hormones are derived from cholesterol They have similar structure. They are made in only a few organs. Adrenal cortex Gonads Skin Placenta Steroids are lipophilic and diffuse easily across membranes, so cells can not store hormones in vesicles. They synthesize as needed. Steroid hormone receptors are in the cytoplasm or in the nucleus. Ultimate destination is nucleus where the complex acts as a transcription factor, binding to DNA and by activationg or repressing one or more genes. Activated genes create mRNA that directs synthesis of new proteins. Any hormone that alters gene activity is said to have a genomic effect on the target cell. When hormones activates genes to direct production of new proteins, there is usually a lag time between hormone-receptor binding and the first biological effects. -

Introduction to the Endocrine System
Introduction to the Endocrine System Hormones 7 Hormones Have Been Known Since Ancient Times What Makes a Chemical a Hormone? Hormones Act by Binding to Receptors Hormone Action Must Be Terminated The Classifi cation of Hormones Most Hormones Are Peptides or Proteins Steroid Hormones Are Derived from Cholesterol Some Hormones Are Derived from Single Amino Acids Control of Hormone Release Hormones Can Be Classifi ed by Their Refl ex Pathways The Endocrine Cell Is the Sensor in the Simplest Endocrine Refl exes Many Endocrine Refl exes Involve the Nervous System Neurohormones Are Secreted into the Blood by Neurons The Pituitary Gland Is Actually Two Fused Glands The Posterior Pituitary Stores and Releases Two Neurohormones The Anterior Pituitary Secretes Six Hormones A Portal System Delivers Hormones from Hypothalamus to Anterior The separation Pituitary of the endocrine Anterior Pituitary Hormones Control Growth, Metabolism, and system into isolated Reproduction subsystems must Feedback Loops Are Diff erent in the Hypothalamic-Pituitary Pathway be recognized as an artifi cial one, Hormone Interactions convenient from In Synergism, the Eff ect of Interacting Hormones Is More Than Additive a pedagogical A Permissive Hormone Allows Another Hormone to Exert Its Full Eff ect point of view but Antagonistic Hormones Have Opposing Eff ects not accurately refl ecting the Endocrine Pathologies interrelated nature Hypersecretion Exaggerates a Hormone’s Eff ects of all these systems. Hyposecretion Diminishes or Eliminates a Hormone’s Eff ects Receptor or Second Messenger Problems Cause Abnormal Tissue — Howard Rasmussen, in Responsiveness Williams’ Textbook of Diagnosis of Endocrine Pathologies Depends on the Complexity of the Endocrinology, 1974 R e fl e x Hormone Evolution Background Basics Focus on . -

Human Anatomy & Physiology the Endocrine System: Part A
10/26/2014 PowerPoint® Lecture Slides Endocrine System: Overview prepared by Human Anatomy Barbara& Physiology Heard, Atlantic Cape Community • Acts with nervous system to coordinate College Ninth Edition and integrate activity of body cells • Influences metabolic activities via C H A P T E R 16 hormones transported in blood • Response slower but longer lasting than nervous system The Endocrine • Endocrinology System: Part A – Study of hormones and endocrine organs © Annie Leibovitz/Contact Press Images © 2013 Pearson Education, Inc. © 2013 Pearson Education, Inc. Endocrine System: Overview Endocrine System: Overview • Controls and integrates • Exocrine glands – Reproduction – Nonhormonal substances (sweat, saliva) – Growth and development – Have ducts to carry secretion to membrane – Maintenance of electrolyte, water, and surface nutrient balance of blood • Endocrine glands – Regulation of cellular metabolism and energy – Produce hormones balance – Lack ducts – Mobilization of body defenses © 2013 Pearson Education, Inc. © 2013 Pearson Education, Inc. Endocrine System: Overview Figure 16.1 Location of selected endocrine organs of the body. Pineal gland Hypothalamus Pituitary gland • Endocrine glands: pituitary, thyroid, Thyroid gland parathyroid, adrenal, and pineal glands Parathyroid glands (on dorsal aspect • Hypothalamus is neuroendocrine organ of thyroid gland) Thymus • Some have exocrine and endocrine functions Adrenal glands – Pancreas, gonads, placenta Pancreas • Other tissues and organs that produce hormones Gonads • Ovary (female) -
Endocrine Glands: Pituitary ,Thyroid ,Parathyroid ,Adrenal ,Pancreas
Endocrine glands: Pituitary ,Thyroid ,Parathyroid ,Adrenal ,Pancreas ,Ovaries & Testes Hormone is a chemical substance released by group of cells to control the function of other type of cells. Target cells refer to cells that contain specific receptors (binding sites) for a particular hormone. Three general classes of hormones: . Proteins and polypeptides -: water soluble – RCs on the cell membrane (anterior and posterior pituitary, pancreas and parathyroid hormones) stored in vesicles until needed . Steroids-: lipid soluble- Derived from cholesterol – RCs in the cytoplasm (adrenal cortex, ovarian and testicular hormones) diffuse across the cell membrane . Derivatives of amino acid tyrosine -: Derived from tyrosine or tryptophan (thyroid hormones lipid soluble + RCs in the nucleus )& (catecholamines water soluble+ RCs on the cell membrane ) Downregulation of hormonal receptors . Increase hormone concentration leads to decrease in the number of active receptors . Most peptide hormones have pulsatile secretion which prevents downregulation Upregulation of hormonal receptors . The hormone induces greater than normal formation of a receptor or intracellular signaling proteins Synergism : Combined action of hormones is more than just additive! Example: Blood glucose levels & synergistic effects of glucagon, cortisol and epinephrine Permissiveness :One hormone allows another hormone to have its full effect Especially during growth Example : Thyroid hormone have permissive effect on growth hormone action. Antagonism :Antagonistic hormones -
Glucocorticoid Metabolism and Function in Ageing Skin
Glucocorticoid Metabolism and Function in Ageing Skin By Ana Tiganescu A thesis presented to the College of Medical and Dental Sciences at the University of Birmingham for the Degree of Doctor of Philosophy Centre for Endocrinology, Diabetes and Metabolism, School of Clinical and Experimental Medicine September 2011 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. SUMMARY The continuing demographic shift towards a greater proportion of elderly individuals in the populations of developing nations and associated increased morbidity is fuelling the drive for increased research into healthy lifespan. Excessive circulating glucocorticoid (GC) levels (e.g. Cushing’s Syndrome) often cause hypertension, osteoporosis, central obesity, muscle weakness, skin thinning and poor wound healing - conditions also commonly associated with the ageing process. Studies in our group investigating the enzyme 11-beta hydroxysteroid dehydrogenase type 1 (11β-HSD1), that generates active GC and regulates their local tissue- specific concentrations, have previously demonstrated increased levels in bone cells derived from older compared to younger donors. This thesis provides exciting new research that reproduces this observation in the organ most noticeably affected by ageing - skin. -
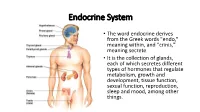
Endocrine System
Endocrine System • The word endocrine derives from the Greek words "endo," meaning within, and "crinis," meaning secrete • It is the collection of glands, each of which secretes different types of hormones that regulate metabolism, growth and development, tissue function, sexual function, reproduction, sleep and mood, among other things. 2006-2007 Nervous System Endocrine System • Communication travels • Chemical messages are in one direction from released into one neuron to another bloodstream or • Has a clear destination extracellular fluid (targets a specific cell) • Target is less specific • Fast acting, short-lived • Hormones are slower- neurotransmitters acting, longer lived (can last days, weeks or years) Endocrine vs. Exocrine Glands • Endocrine Glands – Ductless organs that secrete hormones directly into the bloodstream or extracellular fluid • Exocrine glands – Use ducts (tube-like structures) to transport substances to specific locations inside and outside the body (Ex: sweat glands, mucous glands, salivary glands) Glands • Organ that releases hormones for homeostasis Hormones • Chemical messengers that effect the functions of specifically receptive organs or tissues when transported to them by body fluids • Functions: 1. Regulate growth, development, behavior and reproduction 2. Coordinate production, use, and storage of energy 3. Maintain homeostasis (body temperature, metabolism, excretion, water and salt balance) 4. Response to external stimuli Control of Hormone Release Humoral stimuli - Changing blood levels of ions and nutrients directly stimulates secretion of hormones Neural stimuli- Nerve fibers stimulate hormone release Hormonal stimuli - Hormones stimulate other endocrine organs to release their hormones Chemistry of Hormones • Two main classes 1. Amino acid-based hormones • Amines, thyroxine, peptides, and proteins 2. Steroids • Synthesized from cholesterol • Gonadal and adrenocortical hormones Mechanisms of Hormone Action 1. -
Chapter 11 the Endocrine System Neurotransmitter from a Nerve Cell
Chapter 11 The endocrine system Neurotransmitter from a nerve cell Nerve cell Nerve signals Neurotransmitter molecules Nerve cell / muscle cell / gland cell Hormone from an endocrine cell Hormones are chemical messengers that enter the blood, which carries them from their site of secretion to the cells upon which they act. The endocrine system consists of all those glands, called endocrine glands, that secrete hormones. Secretory vesicles Endocrine cell Hormone molecules Blood vessel Target cell Exocrine gland vs. Endocrine gland Hormone-secreting cells Comparison between nervous system and endocrine system Types of hormones (1) (1) Peptides and proteins (2) Amines (3) Steroids Thyroid hormones Types of hormones (2) Chemical structures of the amine hormones Thyroid hormones By thyroid gland Ps. Adrenal cortex make steroid hormones By adrenal medulla Catecholamines Phenyl-N-methyl transferase (PNMT) Norepinephrine (1X) Epinephrine (4X) By hypothalamus Typical synthesis and secretion of peptide hormones Most hormones @ ribosome @ Golgi @ rER Structures of representative steroid hormones and their structure relationship to cholesterol Adrenal cortex Gonads (testes, ovaries) 1,25-dihydroxyvitamin D (from the kidneys) Steroid synthesis cytochrome P450s The major five hormones secreted from the adrenal cortex Androgens (Testosterone-like hormones) DHEA (1) Organic metabolism Glucocorticoids (2) Responses to stress (3) Regulation of the immune system (1) Na+ and H O retension Mineralocorticoid 2 (2) K+ and H+ excretion in the urine IP3 Angiotensin -

Differential Susceptibility Or Diathesis-Stress
brain sciences Article Differential Susceptibility or Diathesis-Stress: Testing the Moderating Role of Temperament and Cortisol Levels between Fathers’ Parenting and Children’s Aggressive Behavior Eider Pascual-Sagastizabal , Nora del Puerto-Golzarri * and Aitziber Azurmendi Department of Basic Psychological Processes and Their Development, Faculty of Psychology, University of the Basque Country (UPV/EHU), 20018 San Sebastian, Spain; [email protected] (E.P.-S.); [email protected] (A.A.) * Correspondence: [email protected] Abstract: Aggression is a multidimensional behavior that could be caused by different biopsychoso- cial variables. The aim of this study was to explore whether temperament, cortisol and sex moderate the relation between fathers’ parenting style and aggressive behavior in school-aged children, and whether this corresponds to differential susceptibility or diathesis-stress. Participants were 158 chil- dren (88 boys and 70 girls) aged 8 years. Aggressive behavior was measured using the Direct and Indirect Aggression Scale and fathers informed about their child’s temperament and their own par- enting style through the Children’s Behavior Questionnaire and the Parenting Styles and Dimensions Citation: Pascual-Sagastizabal, E.; Questionnaire (respectively). Children’s’ baseline saliva cortisol levels were analyzed through an del Puerto-Golzarri, N.; Azurmendi, enzyme immunoassay technique. The results revealed that high cortisol levels moderated the relation A. Differential Susceptibility or between fathers’ low levels of authoritative parenting and boys’ aggression. Moreover, high negative Diathesis-Stress: Testing the emotionality moderated the relation between permissive paternal parenting and girls’ aggressive Moderating Role of Temperament behavior, with both these relations being consistent with the diathesis-stress theory. and Cortisol Levels between Fathers’ Parenting and Children’s Aggressive Keywords: aggressive behavior; cortisol; temperament; parenting styles; children Behavior.