Combined Comparative Genomics and Gene Expression Analyses Provide Insights Into the Terpene Synthases Inventory in Trichoderma
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Structure-Based Virtual Screening of Hypothetical Inhibitors of the Enzyme Longiborneol Synthase—A Potential Target to Reduce Fusarium Head Blight Disease
J Mol Model (2016) 22: 163 DOI 10.1007/s00894-016-3021-1 ORIGINAL PAPER Structure-based virtual screening of hypothetical inhibitors of the enzyme longiborneol synthase—a potential target to reduce Fusarium head blight disease E. Bresso1 & V. L ero ux 2 & M. Urban3 & K. E. Hammond-Kosack3 & B. Maigret2 & N. F. Martins1 Received: 17 December 2015 /Accepted: 27 May 2016 /Published online: 21 June 2016 # Springer-Verlag Berlin Heidelberg 2016 Abstract Fusarium head blight (FHB) is one of the most compounds from a library of 15,000 drug-like compounds. destructive diseases of wheat and other cereals worldwide. These putative inhibitors of longiborneol synthase provide a During infection, the Fusarium fungi produce mycotoxins that sound starting point for further studies involving molecular represent a high risk to human and animal health. Developing modeling coupled to biochemical experiments. This process small-molecule inhibitors to specifically reduce mycotoxin could eventually lead to the development of novel approaches levels would be highly beneficial since current treatments to reduce mycotoxin contamination in harvested grain. unspecifically target the Fusarium pathogen. Culmorin pos- sesses a well-known important synergistically virulence role Keywords Fusarium mycotoxins . Culmorin . Inhibitors . among mycotoxins, and longiborneol synthase appears to be a Homology modeling . Molecular dynamics . Ensemble key enzyme for its synthesis, thus making longiborneol syn- docking thase a particularly interesting target. This study aims to dis- cover potent and less toxic agrochemicals against FHB. These compounds would hamper culmorin synthesis by inhibiting Introduction longiborneol synthase. In order to select starting molecules for further investigation, we have conducted a structure- Fusarium head blight (FHB), caused by Fusarium based virtual screening investigation. -

Rehmannia Glutinosa-Monocultured Rhizosphere Soil
Comparative Metaproteomic Analysis on Consecutively Rehmannia glutinosa-Monocultured Rhizosphere Soil Linkun Wu1,2, Haibin Wang1,2., Zhixing Zhang1,2., Rui Lin2,3, Zhongyi Zhang1,4, Wenxiong Lin1,2* 1 School of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China, 2 Agroecological Institute, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China, 3 College of Oceanography and Environmental Science, Xiamen University, Xiamen, Fujian, China, 4 Institute of Chinese Medicinal Materials, Henan Agriculture University, Zhengzhou, Henan, China Abstract Background: The consecutive monoculture for most of medicinal plants, such as Rehmannia glutinosa, results in a significant reduction in the yield and quality. There is an urgent need to study for the sustainable development of Chinese herbaceous medicine. Methodology/Principal Findings: Comparative metaproteomics of rhizosphere soil was developed and used to analyze the underlying mechanism of the consecutive monoculture problems of R. glutinosa. The 2D-gel patterns of protein spots for the soil samples showed a strong matrix dependency. Among the spots, 103 spots with high resolution and repeatability were randomly selected and successfully identified by MALDI TOF-TOF MS for a rhizosphere soil metaproteomic profile analysis. These proteins originating from plants and microorganisms play important roles in nutrient cycles and energy flow in rhizospheric soil ecosystem. They function in protein, nucleotide and secondary metabolisms, signal transduction and resistance. Comparative metaproteomics analysis revealed 33 differentially expressed protein spots in rhizosphere soil in response to increasing years of monoculture. Among them, plant proteins related to carbon and nitrogen metabolism and stress response, were mostly up-regulated except a down-regulated protein (glutathione S-transferase) involving detoxification. -
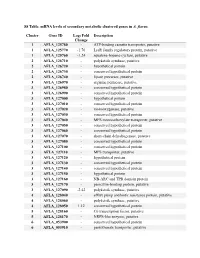
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

Searching for Novel Targets to Control Wheat Head Blight Disease—I-Protein Identification, 3D Modeling and Virtual Screening
Advances in Microbiology, 2016, 6, 811-830 http://www.scirp.org/journal/aim ISSN Online: 2165-3410 ISSN Print: 2165-3402 Searching for Novel Targets to Control Wheat Head Blight Disease—I-Protein Identification, 3D Modeling and Virtual Screening Natália F. Martins1, Emmanuel Bresso1, Roberto C. Togawa1, Martin Urban2, John Antoniw2, Bernard Maigret3, Kim Hammond-Kosack2 1EMBRAPA Recursos Genéticos e Biotecnologia Parque Estação Biológica, Brasília, Brazil 2Department of Plant Biology and Crop Science, Rothamsted Research, Harpenden, UK 3CNRS, LORIA, UMR 7503, Lorraine University, Nancy, France How to cite this paper: Martins, N.F., Abstract Bresso, E., Togawa, R.C., Urban, M., Anto- niw, J., Maigret, B. and Hammond-Kosack, Fusarium head blight (FHB) is a destructive disease of wheat and other cereals. FHB K. (2016) Searching for Novel Targets to occurs in Europe, North America and around the world causing significant losses in Control Wheat Head Blight Disease— production and endangers human and animal health. In this article, we provide the I-Protein Identification, 3D Modeling and strategic steps for the specific target selection for the phytopathogen system wheat- Virtual Screening. Advances in Microbiol- ogy, 6, 811-830. Fusarium graminearum. The economic impact of FHB leads to the need for innova- http://dx.doi.org/10.4236/aim.2016.611079 tion. Currently used fungicides have been shown to be effective over the years, but recently cereal infecting Fusaria have developed resistance. Our work presents a new Received: June 21, 2016 perspective on target selection to allow the development of new fungicides. We de- Accepted: September 11, 2016 veloped an innovative approach combining both genomic analysis and molecular Published: September 14, 2016 modeling to increase the discovery for new chemical compounds with both safety Copyright © 2016 by authors and and low environmental impact. -

Electric Supporting Information (ESI) Crystal Structure and Functional
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2018 Electric Supporting Information (ESI) Crystal structure and functional analysis of large-terpene synthase belonging to a newly found subclass Masahiro Fujihashi,a* Tsutomu Sato,b* Yuma Tanaka,a Daisuke Yamamoto,a Tomoyuki Nishi,b Daijiro Ueda,b Mizuki Murakami,b Yoko Yasuno,c Ai Sekihara,c Kazuma Fuku,c Tetsuro Shinadac & Kunio Mikia* a. Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan. E-mail: [email protected], [email protected] b. Department of Applied Biological Chemistry, Faculty of Agriculture, and Graduate School of Science and Technology, Niigata University, 8050 Ikarashi-2, Niigata 950-2181, Japan. E-mail: [email protected] c. Graduate School of Science, Osaka City University, 3-3-138 Sugimoto, Sumiyoshi, Osaka 558-8585, Japan. S1 Table of Contents Experimental Procedures S3 General procedure S3 Vector construction, expression and purification of BalTS S3 Crystallization and crystallographic analysis of balTS S3 Oligomeric state analysis S4 Analysis of the dimer geometry S4 Vector construction, expression and purification of BsuTS S4 Homology modeling of BsuTS S4 Enzymatic assays S5 Isolation and identification of β-springen S5 Movie Captions S6 Author Contributions S6 Figures Fig. S1 S7 Fig. S2 S8 Fig. S3 S9 Fig. S4 S10 Fig. S5 S17 Fig. S6 S18 Fig. S7, S8 S19 Fig. S9 S20 Fig. S10 S21 Fig. S11 S22 Tables Table S1 S23 Table S2 S24 References S27 S2 Experimental Procedures General procedure NMR spectra were recorded using a Bruker DPX 400 spectrometer at 400 MHz for proton (1H) and 100 MHz for carbon (13C). -

Supporting Information High-Throughput Virtual Screening
Supporting Information High-Throughput Virtual Screening of Proteins using GRID Molecular Interaction Fields Simone Sciabola, Robert V. Stanton, James E. Mills, Maria M. Flocco, Massimo Baroni, Gabriele Cruciani, Francesca Perruccio and Jonathan S. Mason Contents Table S1 S2-S21 Figure S1 S22 * To whom correspondence should be addressed: Simone Sciabola, Pfizer Research Technology Center, Cambridge, 02139 MA, USA Phone: +1-617-551-3327; Fax: +1-617-551-3117; E-mail: [email protected] S1 Table S1. Description of the 990 proteins used as decoy for the Protein Virtual Screening analysis. PDB ID Protein family Molecule Res. (Å) 1n24 ISOMERASE (+)-BORNYL DIPHOSPHATE SYNTHASE 2.3 1g4h HYDROLASE 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE 1.8 1cel HYDROLASE(O-GLYCOSYL) 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I 1.8 1vyf TRANSPORT PROTEIN 14 KDA FATTY ACID BINDING PROTEIN 1.85 1o9f PROTEIN-BINDING 14-3-3-LIKE PROTEIN C 2.7 1t1s OXIDOREDUCTASE 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE 2.4 1t1r OXIDOREDUCTASE 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE 2.3 1q0q OXIDOREDUCTASE 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE 1.9 1jcy LYASE 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE 1.9 1fww LYASE 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE 1.85 1uk7 HYDROLASE 2-HYDROXY-6-OXO-7-METHYLOCTA-2,4-DIENOATE 1.7 1v11 OXIDOREDUCTASE 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT 1.95 1x7w OXIDOREDUCTASE 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT 1.73 1d0l TRANSFERASE 35KD SOLUBLE LYTIC TRANSGLYCOSYLASE 1.97 2bt4 LYASE 3-DEHYDROQUINATE DEHYDRATASE -

Hydroxylation of 1-Deoxypentalenic Acid Mediated by CYP105D7 (SAV 7469) of Streptomyces Avermitilis
The Journal of Antibiotics (2011) 64, 65–71 & 2011 Japan Antibiotics Research Association All rights reserved 0021-8820/11 $32.00 www.nature.com/ja ORIGINAL ARTICLE Pentalenic acid is a shunt metabolite in the biosynthesis of the pentalenolactone family of metabolites: hydroxylation of 1-deoxypentalenic acid mediated by CYP105D7 (SAV_7469) of Streptomyces avermitilis Satoshi Takamatsu1,4, Lian-Hua Xu2,4, Shinya Fushinobu2, Hirofumi Shoun2, Mamoru Komatsu1, David E Cane3 and Haruo Ikeda1 Pentalenic acid (1) has been isolated from many Streptomyces sp. as a co-metabolite of the sesquiterpenoid antibiotic pentalenolactone and related natural products. We have previously reported the identification of a 13.4-kb gene cluster in the genome of Streptomyces avermitilis implicated in the biosynthesis of the pentalenolactone family of metabolites consisting of 13 open reading frames. Detailed molecular genetic and biochemical studies have revealed that at least seven genes are involved in the biosynthesis of the newly discovered metabolites, neopentalenoketolactone, but no gene specifically dedicated to the formation of pentalenic acid (1) was evident in the same gene cluster. The wild-type strain of S. avermitilis, as well as its derivatives, mainly produce pentalenic acid (1), together with neopentalenoketolactone (9). Disruption of the sav7469 gene encoding a cytochrome P450 (CYP105D7), members of which class are associated with the hydroxylation of many structurally different compounds, abolished the production of pentalenic acid (1). The sav7469-deletion mutant derived from SUKA11 carrying pKU462Hptl-clusterDptlH accumulated 1-deoxypentalenic acid (5), but not pentalenic acid (1). Reintroduction of an extra-copy of the sav7469 gene to SUKA11 Dsav7469 carrying pKU462Hptl-clusterDptlH restored the production of pentalenic acid (1). -

X-Ray Fluorescence Analysis Method Röntgenfluoreszenz-Analyseverfahren Procédé D’Analyse Par Rayons X Fluorescents
(19) & (11) EP 2 084 519 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: G01N 23/223 (2006.01) G01T 1/36 (2006.01) 01.08.2012 Bulletin 2012/31 C12Q 1/00 (2006.01) (21) Application number: 07874491.9 (86) International application number: PCT/US2007/021888 (22) Date of filing: 10.10.2007 (87) International publication number: WO 2008/127291 (23.10.2008 Gazette 2008/43) (54) X-RAY FLUORESCENCE ANALYSIS METHOD RÖNTGENFLUORESZENZ-ANALYSEVERFAHREN PROCÉDÉ D’ANALYSE PAR RAYONS X FLUORESCENTS (84) Designated Contracting States: • BURRELL, Anthony, K. AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Los Alamos, NM 87544 (US) HU IE IS IT LI LT LU LV MC MT NL PL PT RO SE SI SK TR (74) Representative: Albrecht, Thomas Kraus & Weisert (30) Priority: 10.10.2006 US 850594 P Patent- und Rechtsanwälte Thomas-Wimmer-Ring 15 (43) Date of publication of application: 80539 München (DE) 05.08.2009 Bulletin 2009/32 (56) References cited: (60) Divisional application: JP-A- 2001 289 802 US-A1- 2003 027 129 12164870.3 US-A1- 2003 027 129 US-A1- 2004 004 183 US-A1- 2004 017 884 US-A1- 2004 017 884 (73) Proprietors: US-A1- 2004 093 526 US-A1- 2004 235 059 • Los Alamos National Security, LLC US-A1- 2004 235 059 US-A1- 2005 011 818 Los Alamos, NM 87545 (US) US-A1- 2005 011 818 US-B1- 6 329 209 • Caldera Pharmaceuticals, INC. US-B2- 6 719 147 Los Alamos, NM 87544 (US) • GOLDIN E M ET AL: "Quantitation of antibody (72) Inventors: binding to cell surface antigens by X-ray • BIRNBAUM, Eva, R. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

Supplementary Materials
Supplementary Materials OH 8 H H8 H C N OH 3 7 3 2 4 H1 1 5 O H4 O 6 H6’ H3 OH H6 H1’ H2 H5 3.9 3.8 3.7 3.6 3.5 3.4 3.3 3.2 ppm 4.0 3.8 3.6 3.4 3.2 3.0 2.8 2.6 2.4 2.2 ppm Figure S1. 1D 1H spectrum of 1-dglcnac and signal assignments ppm 2.0 H8 2.5 OH 8 H H C N OH 3 7 3 2 4 3.0 1 5 H1 O H5 O 6 H4 H3 OH 3.5 H6 H2 H6’ H1’ 4.0 4.0 3.8 3.6 3.4 3.2 3.0 2.8 2.6 2.4 2.2 2.0 ppm Figure S2. 1H-1H TOCSY 2D NMR spectra for 1-dglcnac and signal assignments ppm 3.0 H1 3.2 H5 H4 3.4 H3 3.6 H6 H6’ H2 3.8 H1’ 4.0 4.1 4.0 3.9 3.8 3.7 3.6 3.5 3.4 3.3 3.2 3.1 3.0 ppm Figure S3. 1H-1H COSY 2D NMR spectra for 1-dglcnac and signal assignment ppm -0.02 -0.01 0.00 0.01 0.02 H1’ H6’ H2 H6 H3 H4 H5 H1 3.9 3.8 3.7 3.6 3.5 3.4 3.3 3.2 3.1 ppm Figure S4. 1H-1H JRES 2D NMR spectra for 1-dglcnac and signal assignments ppm 20 C8(22.9) 40 OH 8 H H C N OH C2(53.2) 3 7 3 2 4 60 1 5 C6(63.2) C6(63.2) O 6 C1(69.2) C1(69.2) O C4(72.6) C3(77.0) OH 80 C5(82.7) 100 4.0 3.8 3.6 3.4 3.2 3.0 2.8 2.6 2.4 2.2 2.0 ppm Figure S5. -

Diene Synthase for the Production of Novel Products
Engineering Selina-4(15),7(11)-diene synthase for the Production of Novel Products Emily Turri Submitted in accordance with the requirements for the degree of Doctor of Philosophy The University of Leeds Astbury Centre for Structural Molecular Biology September 2020 The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Emily Turri to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2020 The University of Leeds and Emily Turri 2 Acknowledgements Throughout my PhD I have been extremely lucky to receive support and encouragement from family, friends, colleagues and the university. Without these people and their support, this thesis would not be possible. First, I would like to thank my supervisors Alan Berry and Adam Nelson for their encouragement, guidance and patience over these four years. I would also like to thank Glyn Hemsworth and Nasir Khan for their continued advice and support. I am very grateful to Glyn Hemsworth, Sheena Radford, David Brockwell and John Blacker for letting me use their equipment while offering me direction to give me the best results. In addition, I would like to thank James Ault, Rachel George, Mary Bayana, Mark Howard and Ricardo Labes for their technical assistance and biochemical analysis. My PhD experience would have been nothing without the present and past members of the Hemrry’s and Radford’s with specific thanks to Jess, Alex, Jess and Dan. -

(12) Patent Application Publication (10) Pub. No.: US 2004/0229367 A1 Berka Et Al
US 2004O229367A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2004/0229367 A1 Berka et al. (43) Pub. Date: Nov. 18, 2004 (54) METHODS FOR MONITORING MULTIPLE Related U.S. Application Data GENE EXPRESSION (60) Division of application No. 09/533,559, filed on Mar. (75) Inventors: Randy M. Berka, Davis, CA (US); 22, 2000, which is a continuation-in-part of applica Michael W. Rey, Davis, CA (US); tion No. 09/273,623, filed on Mar. 22, 1999, now Jeffrey R. Shuster, Davis, CA (US); abandoned. Sakari Kauppinen, Smoerum (DK); Ib Groth Clausen, Hillerod (DK); Peter Publication Classification Bjarke Olsen, Copenhagen (DK) 51)1) Int. Cl.C.7 ............................. C12N 15/745/74; C12N 1/16 Correspondence Address: (52) U.S. Cl. ......................................... 435/.484; 435/254.3 NOVOZYMES BIOTECH, INC. (57) ABSTRACT 1445. DREWAVE The present invention relates to methods for monitoring DAVIS, CA 95616 (US) differential expression of a plurality of genes in a first filamentous fungal cell relative to expression of the same genes in one or more Second filamentous fungal cells using (73) Assignees: Novozymes Biotech, Inc., Davis, CA; microarrays containing filamentous fungal expressed Novozymes A/S, Inc., Bagsvaerd (DK) Sequenced tags. The present invention also relates to fila mentous fungal expressed Sequenced tags and to computer (21) Appl. No.: 10/653,047 readable media and Substrates containing Such expressed Sequenced tags for monitoring expression of a plurality of (22) Filed: Aug. 29, 2003 genes in filamentous fungal cells. US 2004/0229367 A1 Nov. 18, 2004 METHODS FOR MONITORING MULTIPLE GENE organisms whose genomes have not been Sequenced.