At Least 23 Genera Instead of One: the Case of Iris L. Sl
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

(Iris Sibirica) Im Westlichen Bodenseegebiet 27-51 ©Staatl
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Carolinea - Beiträge zur naturkundlichen Forschung in Südwestdeutschland Jahr/Year: 2011 Band/Volume: 69 Autor(en)/Author(s): Peintinger Markus Artikel/Article: Verbreitung, Populationsdynamik und Vergesellschaftung der Sibirischen Schwertlilie (Iris sibirica) im westlichen Bodenseegebiet 27-51 ©Staatl. Mus. f. Naturkde Karlsruhe & Naturwiss. Ver. Karlsruhe e.V.; download unter www.zobodat.at carolinea, 69 (2011): 27-51, 4 Abb.; Karlsruhe, 15.12.2011 27 Verbreitung, Populationsdynamik und Vergesellschaftung der Sibirischen Schwertlilie (Iris sibirica) im westlichen Bodenseegebiet MARKUS PEINTINGER Kurzfassung alle Streuwiesen mit Iris sibirica befinden sich in Natur- Die Sibirische Schwertlilie (Iris sibirica) ist eine ty- schutzgebieten und werden aus Naturschutzgründen pische Art der Streuwiesen (Molinion caeruleae) am einmal im Jahr gemäht. Bodenseeufer. Der beobachtete Rückgang einiger Po- pulationen wurde zum Anlass genommen, um Verände- Abstract rungen in der Populationsgröße und der Artenzusam- Distribution, population dynamics and phytoso- mensetzung von Iris sibirica-Wiesen zu untersuchen. ciology of Iris sibirica in the western part of Lake Es wurde hauptsächlich drei Fragen nachgegangen: Constance 1. Wie ist die aktuelle Verbreitung von Iris sibirica und Iris sibirica is a characteristic species for calcareous hat sich der Bestand in den letzten hundert Jahren ver- fen meadows (Molinion caeruleae) at the shore of Lake ändert? 2. Gibt es einen langfristigen Populationstrend Constance. Since a decline of local populations was 1992-2008? 3. Hat sich die Artenzusammensetzung observed, a more detailed study was initiated to exa- der Streuwiesen mit Iris sibirica-Wiesen zwischen den mine changes in population size and species compo- zwei untersuchten Zeitperioden 1988-1993 und 2003- sition of Iris sibirica meadows. -

Oakland Nurseries Inc Blackberry Lily
Blackberry Lily Iris domestica Plant Height: 18 inches Flower Height: 3 feet Spread: 24 inches Sunlight: Hardiness Zone: 4b Other Names: Belamcanda chinensis, Leopard Lily Ornamental Features Blackberry Lily features solitary orange trumpet-shaped flowers with scarlet overtones and red spots at the ends of the stems in early summer. Its sword-like leaves remain green in color throughout the Blackberry Lily flowers season. The black fruits are held in clusters from late summer to early Photo courtesy of NetPS Plant Finder fall. Landscape Attributes Blackberry Lily is an open herbaceous perennial with an upright spreading habit of growth. Its medium texture blends into the garden, but can always be balanced by a couple of finer or coarser plants for an effective composition. This is a relatively low maintenance plant, and is best cleaned up in early spring before it resumes active growth for the season. Gardeners should be aware of the following characteristic(s) that may warrant special consideration; - Spreading Blackberry Lily is recommended for the following landscape applications; - General Garden Use - Container Planting Planting & Growing Blackberry Lily will grow to be about 18 inches tall at maturity extending to 3 feet tall with the flowers, with a spread of 24 inches. It grows at a slow rate, and under ideal conditions can be expected to live for approximately 5 years. Columbus Garden Center - 1156 Oakland Park Avenue, Columbus, OH 43224-3317 Phone: 614-268-3511 Fax: 614-784-7700 Delaware Garden Center - 25 Kilbourne Road, Delaware, OH 43015 Phone: 740-548-6633 Fax: 740-363-2091 Dublin Garden Center - 4261 West Dublin-Granville Road, Dublin, Ohio 43017 Phone: 614-874-2400 Fax: 614-874-2420 New Albany Garden Center - 5211 Johnstown Rd, New Albany, Ohio 43054 Phone: 614-917-1020 Fax: 614-917-1023 This plant should only be grown in full sunlight. -
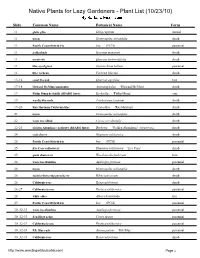
Native Plants for Lazy Gardeners - Plant List (10/23/10)
Native Plants for Lazy Gardeners - Plant List (10/23/10) Slide Common Name Botanical Name Form 11 globe gilia Gilia capitata annual 11 toyon Heteromeles arbutifolia shrub 11 Pacific Coast Hybrid iris Iris (PCH) perennial 11 goldenbush Isocoma menziesii shrub 11 scrub oak Quercus berberidifolia shrub 11 blue-eyed grass Sisyrinchium bellum perennial 11 lilac verbena Verbena lilacina shrub 13-16 coast live oak Quercus agrifolia tree 17-18 Howard McMinn man anita Arctostaphylos 'Howard McMinn' shrub 19 Philip Mun keckiella (RSABG Intro) Keckiella 'Philip Munz' ine 19 woolly bluecurls Trichostema lanatum shrub 19-20 Ray Hartman California lilac Ceanothus 'Ray Hartman' shrub 21 toyon Heteromeles arbutifolia shrub 22 western redbud Cercis occidentalis shrub 22-23 Golden Abundance barberry (RSABG Intro) Berberis 'Golden Abundance' (MAHONIA) shrub 2, coffeeberry Rhamnus californica shrub 25 Pacific Coast Hybrid iris Iris (PCH) perennial 25 Eve Case coffeeberry Rhamnus californica '. e Case' shrub 25 giant chain fern Woodwardia fimbriata fern 26 western columbine Aquilegia formosa perennial 26 toyon Heteromeles arbutifolia shrub 26 fuchsia-flowering gooseberry Ribes speciosum shrub 26 California rose Rosa californica shrub 26-27 California fescue Festuca californica perennial 28 white alder Alnus rhombifolia tree 29 Pacific Coast Hybrid iris Iris (PCH) perennial 30 032-33 western columbine Aquilegia formosa perennial 30 032-33 San Diego sedge Carex spissa perennial 30 032-33 California fescue Festuca californica perennial 30 032-33 Elk Blue rush Juncus patens '.l1 2lue' perennial 30 032-33 California rose Rosa californica shrub http://www weedingwildsuburbia com/ Page 1 30 032-3, toyon Heteromeles arbutifolia shrub 30 032-3, fuchsia-flowering gooseberry Ribes speciosum shrub 30 032-3, Claremont pink-flowering currant (RSA Intro) Ribes sanguineum ar. -

Philip J. Savage, Jr
ISSUE 73 M4SNOU4 Philip J. Savage, Jr. May 8, 1917 — October 13, 2002 Phil Savnge, /r. , an icon m the Magnolia worhl, passed au ay last October of conr plica- lions from the West Nile virus. Phil, a renotvrred magnolin Irybcidrzer, specialized in breeding mngnolias that u&cre cold Irardy. Becnuse of this, nragnolia entirusiasts living in colder climalcs now have morry rrrorc choices thnn did Dennis Ledvh rn u&hen he bought Iris lrouse irr Green Bay, Wiscorrsin in thc Intr typos Plril hns left an errduring legaclt n&itlr tire mnny /Inc, cold-lurrdy hybrids Ire bred. Following arc set&eral rwni niscclrces fronr irrdividuals who toere deqviy in/luencerl by Phil mrd Iris u&ork u&ith mngrrolins. DENNIs LEDVINA WIUTES. Back in the late yos the landscaping around my new house consisted of three magnolias: two M. x sorrlarrgeana, and a M. x loebneri 'MerrilL' At the time, these were the only magnolias gener- ally available at local nurseries. As I watched these magnolias bloom each spring, I became more intrigued with their beautiful Bowers and began driv- ing around Green Bay to observe and admire some of the established trees. My admiration for the genus contin- ued to grow each year as I began col- lecting more information about these magnificent plants. One summer I was in the Detroit area and I decided to call this magnolia expert, Phil Savage, that I had read so much about. I can vividly remember calling Phil from a telephone booth on Telegraph Road and finding mysell, an unknown amateur, talking to a magnolia expert who from the first made me feel like a lifelong friend. -

AGS Seed List No 69 2020
Seed list No 69 2020-21 Garden Collected Seed 1001 Abelia floribunda 1057 Agrostemma githago 1002 Abies koreana 1058 Albuca canadensis (L. -

May 15, 2016 Passing Peony and Iris Plants on from Generation to Generation Annette Meyer Heisdorffer Daviess County Extension Agent for Horticulture
May 15, 2016 Passing Peony and Iris Plants on from Generation to Generation Annette Meyer Heisdorffer Daviess County Extension Agent for Horticulture After lunch on Mother’s Day, my mom and I surveyed her garden, especially the peonies. We both agreed that I needed to propagate her peonies and plant them in my garden. These are special, because I remember them growing in my grandmother’s garden. Peonies are commonly passed down from generation to generation. My goal is to someday share them with my twins. Our discussion included the irises, which are another heritage plant. Both plants are blooming beautifully in May and are spectacular in the garden. Information about these two plants will be provided in this article. Peony (Paeonia officinalis, Paeonia lactiflora, and hybrids) is a herbaceous perennial, which means at the end of the growing season it will die back to the ground. However, the plant returns year after year. Peonies grow best in full sun and well-drained soil. There are tree peonies (Paeonia suffruticosa) which have a woody stem, but those are not as common and require different growing conditions. The tree peony will not be discussed here. According to Dr. Rick Durham, Extension Specialist for Consumer Horticulture, peonies can be found in landscapes across Kentucky. Peonies have a long life span and are commonly grown in the garden. When planting the root, make sure it is not too deep. The eyes or bud should be just below the surface of the soil. If it is planted too deeply, the plants won't bloom. -

Iris for the Home Gardener a Rainbow of Colors in Many Shapes and Sizes Bob Lyons
Iris for the Home Gardener A Rainbow of Colors in Many Shapes and Sizes Bob Lyons FEW PLANTS HAVE AS MUCH HISTORY and affection among gardeners than iris. In Greek mythology, Iris is the personification of the rainbow and messenger of the Gods, and indeed, Iris appear in many magical colors—a large and diverse genus. Some have large showy flowers, others more I. ‘Black Gamecock’ understated; some grow in clumps, others spread; some prefer I. ensata ‘Angelic Choir’ it dry, others are more partial to moist, even wet conditions; and some grow from bulbs, while others return each year from rhizomes just beneath the soil surface. How does one tell them apart and make the right choice for a home garden? Fortunately, horticulturists and iris enthusiasts have developed a system of organization to make sense out of the vast world of irises. Three groups that account for more than 75% of the commercial iris market today are the Bearded Iris, Siberian Iris, and Japanese Iris. Each group recognizes the best of the best with prestigious national awards, noted in the descriptions that follow. The Dykes Medal is awarded to the finest iris of any class. More iris plants are described in the “Plant Descriptions: I. ×pseudata ‘Aichi no Kagayaki’ I. ensata ‘Cascade Crest’ Perennial” section. Latin Name Common Name Mature Size Light Soil Pot Size Price Iris ‘Black Gamecock’ Louisiana Iris 2–3 .8 d 1 g $14 Late; stunning blue black, velvet-colored flowers; hummingbird haven; can grow in 4 inches of standing water; DeBaillon Medal. Iris ×pseudata ‘Aichi no Kagayaki’ Iris Hybrid 2 . -

Fall 2013 NARGS
Rock Garden uar terly � Fall 2013 NARGS to ADVERtISE IN thE QuARtERly CoNtACt [email protected] Let me know what yo think A recent issue of a chapter newsletter had an item entitled “News from NARGS”. There were comments on various issues related to the new NARGS website, not all complimentary, and then it turned to the Quarterly online and raised some points about which I would be very pleased to have your views. “The good news is that all the Quarterlies are online and can easily be dowloaded. The older issues are easy to read except for some rather pale type but this may be the result of scanning. There is amazing information in these older issues. The last three years of the Quarterly are also online but you must be a member to read them. These last issues are on Allen Press’s BrightCopy and I find them harder to read than a pdf file. Also the last issue of the Quarterly has 60 extra pages only available online. Personally I find this objectionable as I prefer all my content in a printed bulletin.” This raises two points: Readability of BrightCopy issues versus PDF issues Do you find the BrightCopy issues as good as the PDF issues? Inclusion of extra material in online editions only. Do you object to having extra material in the online edition which can not be included in the printed edition? Please take a moment to email me with your views Malcolm McGregor <[email protected]> CONTRIBUTORS All illustrations are by the authors of articles unless otherwise stated. -

Karyological Studies on Iris Japonica Thunb. and Its Allies1) 2) by K
180 Cytologia 10 Karyological Studies on Iris japonica Thunb. and Its Allies1) 2) By K. Yasui Tokyo Imperial University (With 8 text figures) Receic ed MTV 28, 1939 Introduction There are several karyological investigations on Iris japonica. KAZAO (1928) considered the sporophyte of this species as an auto triploid having 54 chromosomes. But SIMONET (1932, 1934) re ported I. japonica as a diploid plant with 34 chromosomes in its root-tip cells, and considered I. japonica var aphrodite as a hyper triploid and interpreted the chromosome constitution as 2n=51+3, the latter 3 chromosomes being considered as derived from 3 of 51 chromosomes in I. japonica by fragmentation which occurred att their median constrictions. The karyological investigation in I. japonica and the other 2 allied species, which were kindly put at my deposal by Dr. S. IMA MURA of Kyoto Imp. University, gave me some results different from those of the previous authors as is described below. Material and Method In the fixing of the root-tip cells both NAVASHIN's and LEWITSKY's solutions were used. When fixed in the latter, the chromosomes were found longer and slender than when fixed in the former solution, but there was no essential difference in the distribu tion of chromosomes in the equatorial plate. NEWTON'S gentian violet staining was generally adopted. Materials for the study of meiosis were fixed mostly with NAVASHIN'S fluid and stained with HEIDENHAIN'S iron-alum haematoxylin. Acetocarmine smears were also tried in the study of the pollen mother cells. Drawings were made with a camera lucida. -
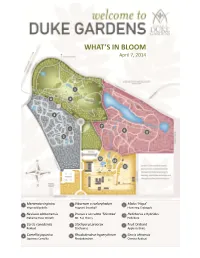
What's in Bloom
WHAT’S IN BLOOM April 7, 2014 5 4 6 2 7 1 9 8 3 12 10 11 1 Mertensia virginica 5 Viburnum x carlcephalum 9 Malus ‘Hopa’ Virginia Bluebells Fragrant Snowball Flowering Crabapple 2 Neviusia alabamensis 6 Prunus x serrulata ‘Shirotae’ 10 Helleborus x hybridus Alabama Snow Wreath Mt. Fuji Cherry Hellebore 3 Cercis canadensis 7 Stachyurus praecox 11 Fruit Orchard Redbud Stachyurus Apple cultivars 4 Camellia japonica 8 Rhododendron hyperythrum 12 Cercis chinensis Japanese Camellia Rhododendron Chinese Redbud WHAT’S IN BLOOM April 7, 2014 BLOMQUIST GARDEN OF NATIVE PLANTS Amelanchier arborea Common Serviceberry Sanguinaria canadensis Bloodroot Cornus florida Flowering Dogwood Stylophorum diphyllum Celandine Poppy Thalictrum thalictroides Rue Anemone Fothergilla major Fothergilla Trillium decipiens Chattahoochee River Trillium Hepatica nobilis Hepatica Trillium grandiflorum White Trillium Hexastylis virginica Wild Ginger Hexastylis minor Wild Ginger Trillium pusillum Dwarf Wakerobin Illicium floridanum Florida Anise Tree Trillium stamineum Blue Ridge Wakerobin Malus coronaria Sweet Crabapple Uvularia sessilifolia Sessileleaf Bellwort Mertensia virginica Virginia Bluebells Pachysandra procumbens Allegheny spurge Prunus americana American Plum DORIS DUKE CENTER GARDENS Camellia japonica Japanese Camellia Pulmonaria ‘Diana Clare’ Lungwort Cercis canadensis Redbud Prunus persica Flowering Peach Puschkinia scilloides Striped Squill Cercis chinensis Redbud Sanguinaria canadensis Bloodroot Clematis armandii Evergreen Clematis Spiraea prunifolia Bridalwreath -

These De Doctorat De L'universite Paris-Saclay
NNT : 2016SACLS250 THESE DE DOCTORAT DE L’UNIVERSITE PARIS-SACLAY, préparée à l’Université Paris-Sud ÉCOLE DOCTORALE N° 567 Sciences du Végétal : du Gène à l’Ecosystème Spécialité de doctorat (Biologie) Par Mlle Nour Abdel Samad Titre de la thèse (CARACTERISATION GENETIQUE DU GENRE IRIS EVOLUANT DANS LA MEDITERRANEE ORIENTALE) Thèse présentée et soutenue à « Beyrouth », le « 21/09/2016 » : Composition du Jury : M., Tohmé, Georges CNRS (Liban) Président Mme, Garnatje, Teresa Institut Botànic de Barcelona (Espagne) Rapporteur M., Bacchetta, Gianluigi Università degli Studi di Cagliari (Italie) Rapporteur Mme, Nadot, Sophie Université Paris-Sud (France) Examinateur Mlle, El Chamy, Laure Université Saint-Joseph (Liban) Examinateur Mme, Siljak-Yakovlev, Sonja Université Paris-Sud (France) Directeur de thèse Mme, Bou Dagher-Kharrat, Magda Université Saint-Joseph (Liban) Co-directeur de thèse UNIVERSITE SAINT-JOSEPH FACULTE DES SCIENCES THESE DE DOCTORAT DISCIPLINE : Sciences de la vie SPÉCIALITÉ : Biologie de la conservation Sujet de la thèse : Caractérisation génétique du genre Iris évoluant dans la Méditerranée Orientale. Présentée par : Nour ABDEL SAMAD Pour obtenir le grade de DOCTEUR ÈS SCIENCES Soutenue le 21/09/2016 Devant le jury composé de : Dr. Georges TOHME Président Dr. Teresa GARNATJE Rapporteur Dr. Gianluigi BACCHETTA Rapporteur Dr. Sophie NADOT Examinateur Dr. Laure EL CHAMY Examinateur Dr. Sonja SILJAK-YAKOVLEV Directeur de thèse Dr. Magda BOU DAGHER KHARRAT Directeur de thèse Titre : Caractérisation Génétique du Genre Iris évoluant dans la Méditerranée Orientale. Mots clés : Iris, Oncocyclus, région Est-Méditerranéenne, relations phylogénétiques, status taxonomique. Résumé : Le genre Iris appartient à la famille des L’approche scientifique est basée sur de nombreux Iridacées, il comprend plus de 280 espèces distribuées outils moléculaires et génétiques tels que : l’analyse de à travers l’hémisphère Nord. -

Potted Sale Plant MASTER LIST May 11.Xlsx
9/7/2020 Texas Discovery Gardens Plant Sale List Page 1 of 16 ALPHABETICAL BY PLANT GROUP** Tx=Tx Pollinators Sun Req. native Common Name Botanic Name Height Plant Group Plant Type Host / Attracted X=Not Nectar Full sun X-E Af Whistling Thorn Acacia (now Heat & Drought Tolerant Deciduous Vachellia) drepanolobium Hot sun X-Mex Truncate Parry's Agave parryi v. 3' Heat & Drought Tolerant Evergreen Agave truncata Hot sun X-Mex Green Giant Agave salmiana 10' Heat & Drought Tolerant Min. 30°F. Agave Full to part X Trunking Beschorneria 3-5' Heat & Drought Tolerant Min. 30°F. N sun Beschorneria albiflora Hot sun Tx Cholla Cactus Cylindropuntia Heat & Drought Tolerant Evergreen imbricata Hot sun Tx Smooth-leaf Sotol Dasylirion 3-4' Heat & Drought Tolerant Evergreen leiophyllum Hot sun Tx Texas Sotol Dasylirion texanum 4' to 5' Heat & Drought Tolerant Evergreen N Hot sun Tx Barrel Cactus Ferocactus wislizeni 3 to 6 ft Heat & Drought Tolerant Perennial N Hot sun Tx Giant Hesperaloe Hesperaloe funifera 6' X 6' Heat & Drought Tolerant Evergreen N Hummingbirds Full to part X-Mex Night Blooming Hesperaloe 5' X 6' Heat & Drought Tolerant Evergreen N Hummingbirds, sun Hesperaloe nocturna moths Hot sun Tx Red Yucca Hesperaloe 4' Heat & Drought Tolerant Evergreen N Hummingbirds parviflora Hot sun Tx Yellow Yucca Hesperaloe 4' X 4' Heat & Drought Tolerant Evergreen N Hummingbirds parviflora yellow Full to part Tx Devil's Shoestring Nolina 3' X 3' Heat & Drought Tolerant Evergreen N sun lindheimeriana Part sun Tx Texas Beargrass Nolina texana Heat & Drought