Short Methionine Synthases Morgan N
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Methionine Synthase Supports Tumor Tetrahydrofolate Pools
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.05.284521; this version posted September 7, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Methionine synthase supports tumor tetrahydrofolate pools Joshua Z. Wang1,2,#, Jonathan M. Ghergurovich1,3,#, Lifeng Yang1,2, and Joshua D. Rabinowitz1,2,* 1 Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, New Jersey, USA 2 Department of Chemistry, Princeton University, Princeton, New Jersey, USA 3 Department of Molecular Biology, Princeton University, Princeton, New Jersey, USA # These authors contributed equally to this work. *Corresponding author: Joshua Rabinowitz Department of Chemistry and the Lewis-Sigler Institute for Integrative Genomics, Princeton University, Washington Rd, Princeton, NJ 08544, USA Phone: (609) 258-8985; e-mail: [email protected] Conflict of Interest Disclosure: J.D.R. is a paid advisor and stockholder in Kadmon Pharmaceuticals, L.E.A.F. Pharmaceuticals, and Rafael Pharmaceuticals; a paid consultant of Pfizer; a founder, director, and stockholder of Farber Partners and Serien Therapeutics. JDR and JMG are inventors of patents in the area of folate metabolism held by Princeton University. 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.05.284521; this version posted September 7, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Mammalian cells require activated folates to generate nucleotides for growth and division. The most abundant circulating folate species is 5-methyl tetrahydrofolate (5-methyl- THF), which is used to synthesize methionine from homocysteine via the cobalamin-dependent enzyme methionine synthase (MTR). -

Defining Sources of Nutrient Limitation for Tumors By
Defining sources of nutrient limitation for tumors by Mark Robert Sullivan B.S. Molecular Biology and Chemistry University of Pittsburgh (2013) Submitted to the Department of Biology in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY at the MASSACHUSETTS INSTITUTE OF TECHNOLOGY June 2019 © 2019 Mark R. Sullivan. All rights reserved. The author hereby grants to MIT permission to reproduce and to distribute publicly paper and electronic copies of this thesis document in whole or in part in any medium now known or hereafter created. Signature of Author.......................................................................................................................... Department of Biology April 30, 2019 Certified by ...................................................................................................................................... Matthew G. Vander Heiden Associate Professor of Biology Thesis Supervisor Accepted by...................................................................................................................................... Amy E. Keating Professor of Biology Co-Director, Biology Graduate Committee 1 2 Defining sources of nutrient limitation for tumors by Mark Robert Sullivan Submitted to the Department of Biology on April 30, 2019 in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in Biology ABSTRACT Tumor growth requires that cancer cells accumulate sufficient biomass to grow and divide. To accomplish this, tumor cells must acquire -

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -
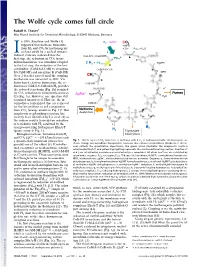
The Wolfe Cycle Comes Full Circle
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Involvements of Hyperhomocysteinemia in Neurological Disorders
H OH metabolites OH Review Involvements of Hyperhomocysteinemia in Neurological Disorders Marika Cordaro 1,† , Rosalba Siracusa 2,† , Roberta Fusco 2 , Salvatore Cuzzocrea 2,3,* , Rosanna Di Paola 2,* and Daniela Impellizzeri 2 1 Department of Biomedical, Dental and Morphological and Functional Imaging, University of Messina, Via Consolare Valeria, 98125 Messina, Italy; [email protected] 2 Department of Chemical, Biological, Pharmaceutical and Environmental Sciences, University of Messina, 98166 Messina, Italy; [email protected] (R.S.); [email protected] (R.F.); [email protected] (D.I.) 3 Department of Pharmacological and Physiological Science, Saint Louis University School of Medicine, Saint Louis, MO 63104, USA * Correspondence: [email protected] (S.C.); [email protected] (R.D.P.); Tel.: +39-090-6765208 (S.C. & R.D.P.) † The authors equally contributed to the review. Abstract: Homocysteine (HCY), a physiological amino acid formed when proteins break down, leads to a pathological condition called hyperhomocysteinemia (HHCY), when it is over a definite limit. It is well known that an increase in HCY levels in blood, can contribute to arterial damage and several cardiovascular disease, but the knowledge about the relationship between HCY and brain disorders is very poor. Recent studies demonstrated that an alteration in HCY metabolism or a deficiency in folate or vitamin B12 can cause altered methylation and/or redox potentials, that leads to a modification on calcium influx in cells, or into an accumulation in amyloid and/or tau protein involving a cascade of events that culminate in apoptosis, and, in the worst conditions, neuronal death. The present review will thus summarize how much is known about the possible role of HHCY in neurodegenerative disease. -

Annotation Guidelines for Experimental Procedures
Annotation Guidelines for Experimental Procedures Developed By Mohammed Alliheedi Robert Mercer Version 1 April 14th, 2018 1- Introduction and background information What is rhetorical move? A rhetorical move can be defined as a text fragment that conveys a distinct communicative goal, in other words, a sentence that implies an author’s specific purpose to readers. What are the types of rhetorical moves? There are several types of rhetorical moves. However, we are interested in 4 rhetorical moves that are common in the method section of a scientific article that follows the Introduction Methods Results and Discussion (IMRaD) structure. 1- Description of a method: It is concerned with a sentence(s) that describes experimental events (e.g., “Beads with bound proteins were washed six times (for 10 min under rotation at 4°C) with pulldown buffer and proteins harvested in SDS-sample buffer, separated by SDS-PAGE, and analyzed by autoradiography.” (Ester & Uetz, 2008)). 2- Appeal to authority: It is concerned with a sentence(s) that discusses the use of standard methods, protocols, and procedures. There are two types of this move: - A reference to a well-established “standard” method (e.g., the use of a method like “PCR” or “electrophoresis”). - A reference to a method that was previously described in the literature (e.g., “Protein was determined using fluorescamine assay [41].” (Larsen, Frandesn and Treiman, 2001)). 3- Source of materials: It is concerned with a sentence(s) that lists the source of biological materials that are used in the experiment (e.g., “All microalgal strains used in this study are available at the Elizabeth Aidar Microalgae Culture Collection, Department of Marine Biology, Federal Fluminense University, Brazil.” (Larsen, Frandesn and Treiman, 2001)). -

Untargetted Metabolomic Exploration of the Mycobacterium Tuberculosis Stress Response to Cinnamon Essential Oil
biomolecules Article Untargetted Metabolomic Exploration of the Mycobacterium tuberculosis Stress Response to Cinnamon Essential Oil Elwira Sieniawska 1,* , Rafał Sawicki 2 , Joanna Golus 2 and Milen I. Georgiev 3,4 1 Chair and Department of Pharmacognosy, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland 2 Chair and Department of Biochemistry and Biotechnology, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland; [email protected] (R.S.); [email protected] (J.G.) 3 Group of Plant Cell Biotechnology and Metabolomics, The Stephan Angeloff Institute of Microbiology, Bulgarian Academy of Sciences, 139 Ruski Blvd., 4000 Plovdiv, Bulgaria; [email protected] 4 Center of Plant Systems Biology and Biotechnology, 4000 Plovdiv, Bulgaria * Correspondence: [email protected] Received: 6 January 2020; Accepted: 24 February 2020; Published: 26 February 2020 Abstract: The antimycobacterial activity of cinnamaldehyde has already been proven for laboratory strains and for clinical isolates. What is more, cinnamaldehyde was shown to threaten the mycobacterial plasma membrane integrity and to activate the stress response system. Following promising applications of metabolomics in drug discovery and development we aimed to explore the mycobacteria response to cinnamaldehyde within cinnamon essential oil treatment by untargeted liquid chromatography–mass spectrometry. The use of predictive metabolite pathway analysis and description of produced lipids enabled the evaluation of the stress symptoms shown by bacteria. This study suggests that bacteria exposed to cinnamaldehyde could reorganize their outer membrane as a physical barrier against stress factors. They probably lowered cell wall permeability and inner membrane fluidity, and possibly redirected carbon flow to store energy in triacylglycerols. Being a reactive compound, cinnamaldehyde may also contribute to disturbances in bacteria redox homeostasis and detoxification mechanisms. -

Human Catechol O-Methyltransferase Genetic Variation
Molecular Psychiatry (2004) 9, 151–160 & 2004 Nature Publishing Group All rights reserved 1359-4184/04 $25.00 www.nature.com/mp ORIGINAL RESEARCH ARTICLE Human catechol O-methyltransferase genetic variation: gene resequencing and functional characterization of variant allozymes AJ Shield1, BA Thomae1, BW Eckloff2, ED Wieben2 and RM Weinshilboum1 1Department of Molecular Pharmacology and Experimental Therapeutics, Mayo Medical School, Mayo Clinic, Mayo Foundation, Rochester, MN, USA; 2Department of Biochemistry and Molecular Biology, Mayo Medical School, Mayo Clinic, Mayo Foundation, Rochester, MN, USA Catechol O-methyltransferase (COMT) plays an important role in the metabolism of catecholamines, catecholestrogens and catechol drugs. A common COMT G472A genetic polymorphism (Val108/158Met) that was identified previously is associated with decreased levels of enzyme activity and has been implicated as a possible risk factor for neuropsychiatric disease. We set out to ‘resequence’ the human COMT gene using DNA samples from 60 African-American and 60 Caucasian-American subjects. A total of 23 single nucleotide polymorphisms (SNPs), including a novel nonsynonymous cSNP present only in DNA from African-American subjects, and one insertion/deletion were observed. The wild type (WT) and two variant allozymes, Thr52 and Met108, were transiently expressed in COS-1 and HEK293 cells. There was no significant change in level of COMT activity for the Thr52 variant allozyme, but there was a 40% decrease in the level of activity in cells transfected with the Met108 construct. Apparent Km values of the WT and variant allozymes for the two reaction cosubstrates differed slightly, but significantly, for 3,4-dihydroxybenzoic acid but not for S-adenosyl-L-methionine. -

Recycling of Vitamin B12 and NAD+ Within the Pdu Microcompartment of Salmonella Enterica Shouqiang Cheng Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2010 Recycling of vitamin B12 and NAD+ within the Pdu microcompartment of Salmonella enterica Shouqiang Cheng Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Biochemistry, Biophysics, and Structural Biology Commons Recommended Citation Cheng, Shouqiang, "Recycling of vitamin B12 and NAD+ within the Pdu microcompartment of Salmonella enterica" (2010). Graduate Theses and Dissertations. 11713. https://lib.dr.iastate.edu/etd/11713 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. + Recycling of vitamin B12 and NAD within the Pdu microcompartment of Salmonella enterica by Shouqiang Cheng A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Biochemistry Program of Study Committee: Thomas A. Bobik, Major Professor Alan DiSpirito Basil Nikolau Reuben Peters Gregory J. Phillips Iowa State University Ames, Iowa 2010 Copyright © Shouqiang Cheng, 2010. All rights reserved. ii Table of contents Abstract............................................................................................................................. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

EZH2 in Normal Hematopoiesis and Hematological Malignancies
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 7, No. 3 EZH2 in normal hematopoiesis and hematological malignancies Laurie Herviou2, Giacomo Cavalli2, Guillaume Cartron3,4, Bernard Klein1,2,3 and Jérôme Moreaux1,2,3 1 Department of Biological Hematology, CHU Montpellier, Montpellier, France 2 Institute of Human Genetics, CNRS UPR1142, Montpellier, France 3 University of Montpellier 1, UFR de Médecine, Montpellier, France 4 Department of Clinical Hematology, CHU Montpellier, Montpellier, France Correspondence to: Jérôme Moreaux, email: [email protected] Keywords: hematological malignancies, EZH2, Polycomb complex, therapeutic target Received: August 07, 2015 Accepted: October 14, 2015 Published: October 20, 2015 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Enhancer of zeste homolog 2 (EZH2), the catalytic subunit of the Polycomb repressive complex 2, inhibits gene expression through methylation on lysine 27 of histone H3. EZH2 regulates normal hematopoietic stem cell self-renewal and differentiation. EZH2 also controls normal B cell differentiation. EZH2 deregulation has been described in many cancer types including hematological malignancies. Specific small molecules have been recently developed to exploit the oncogenic addiction of tumor cells to EZH2. Their therapeutic potential is currently under evaluation. This review summarizes the roles of EZH2 in normal and pathologic hematological processes and recent advances in the development of EZH2 inhibitors for the personalized treatment of patients with hematological malignancies. PHYSIOLOGICAL FUNCTIONS OF EZH2 state through tri-methylation of lysine 27 on histone H3 (H3K27me3) [6]. -

Polymorphisms in Methionine Synthase, Methionine Synthase Reductase and Serine Hydroxymethyltransferase, Folate and Alcohol Intake, and Colon Cancer Risk Susan E
University of South Carolina Scholar Commons Faculty Publications Epidemiology and Biostatistics 2008 Polymorphisms in Methionine Synthase, Methionine Synthase Reductase and Serine Hydroxymethyltransferase, Folate and Alcohol Intake, and Colon Cancer Risk Susan E. Steck University of South Carolina - Columbia, [email protected] Temitope O. Keku Lesley M. Butler Joseph Galanko Beri Massa See next page for additional authors Follow this and additional works at: https://scholarcommons.sc.edu/ sph_epidemiology_biostatistics_facpub Part of the Public Health Commons Publication Info Postprint version. Published in Journal of Nutrigenetics and Nutrigenomics, Volume 1, Issue 4, 2008, pages 196-204. Steck, S. E., Keku, T., Butler, L. M., Galanko, J., Massa, B., Millikan, R. C., & Sandler, R. S. (2008). Polymorphisms in methionine synthase, methionine synthase reductase and serine hydroxymethyltransferase, folate and alcohol intake, and colon cancer risk. Journal of Nutrigenetics and Nutrigenomics, 1(4), 196-204. DOI: 10.1159/000136651 © Journal of Nutrigenetics and Nutrigenomics, 2008, Karger http://content.karger.com/produktedb/produkte.asp?DOI=10.1159/000136651 This Article is brought to you by the Epidemiology and Biostatistics at Scholar Commons. It has been accepted for inclusion in Faculty Publications by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Author(s) Susan E. Steck, Temitope O. Keku, Lesley M. Butler, Joseph Galanko, Beri Massa, Robert C. Millikan, and Robert S. Sandler This article is available at Scholar Commons: https://scholarcommons.sc.edu/sph_epidemiology_biostatistics_facpub/303 NIH Public Access Author Manuscript J Nutrigenet Nutrigenomics. Author manuscript; available in PMC 2009 October 7. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: J Nutrigenet Nutrigenomics.