An Epiblast Stem Cell Derived Multipotent Progenitor Population
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

3 Embryology and Development
BIOL 6505 − INTRODUCTION TO FETAL MEDICINE 3. EMBRYOLOGY AND DEVELOPMENT Arlet G. Kurkchubasche, M.D. INTRODUCTION Embryology – the field of study that pertains to the developing organism/human Basic embryology –usually taught in the chronologic sequence of events. These events are the basis for understanding the congenital anomalies that we encounter in the fetus, and help explain the relationships to other organ system concerns. Below is a synopsis of some of the critical steps in embryogenesis from the anatomic rather than molecular basis. These concepts will be more intuitive and evident in conjunction with diagrams and animated sequences. This text is a synopsis of material provided in Langman’s Medical Embryology, 9th ed. First week – ovulation to fertilization to implantation Fertilization restores 1) the diploid number of chromosomes, 2) determines the chromosomal sex and 3) initiates cleavage. Cleavage of the fertilized ovum results in mitotic divisions generating blastomeres that form a 16-cell morula. The dense morula develops a central cavity and now forms the blastocyst, which restructures into 2 components. The inner cell mass forms the embryoblast and outer cell mass the trophoblast. Consequences for fetal management: Variances in cleavage, i.e. splitting of the zygote at various stages/locations - leads to monozygotic twinning with various relationships of the fetal membranes. Cleavage at later weeks will lead to conjoined twinning. Second week: the week of twos – marked by bilaminar germ disc formation. Commences with blastocyst partially embedded in endometrial stroma Trophoblast forms – 1) cytotrophoblast – mitotic cells that coalesce to form 2) syncytiotrophoblast – erodes into maternal tissues, forms lacunae which are critical to development of the uteroplacental circulation. -

Understanding Paraxial Mesoderm Development and Sclerotome Specification for Skeletal Repair Shoichiro Tani 1,2, Ung-Il Chung2,3, Shinsuke Ohba4 and Hironori Hojo2,3
Tani et al. Experimental & Molecular Medicine (2020) 52:1166–1177 https://doi.org/10.1038/s12276-020-0482-1 Experimental & Molecular Medicine REVIEW ARTICLE Open Access Understanding paraxial mesoderm development and sclerotome specification for skeletal repair Shoichiro Tani 1,2, Ung-il Chung2,3, Shinsuke Ohba4 and Hironori Hojo2,3 Abstract Pluripotent stem cells (PSCs) are attractive regenerative therapy tools for skeletal tissues. However, a deep understanding of skeletal development is required in order to model this development with PSCs, and for the application of PSCs in clinical settings. Skeletal tissues originate from three types of cell populations: the paraxial mesoderm, lateral plate mesoderm, and neural crest. The paraxial mesoderm gives rise to the sclerotome mainly through somitogenesis. In this process, key developmental processes, including initiation of the segmentation clock, formation of the determination front, and the mesenchymal–epithelial transition, are sequentially coordinated. The sclerotome further forms vertebral columns and contributes to various other tissues, such as tendons, vessels (including the dorsal aorta), and even meninges. To understand the molecular mechanisms underlying these developmental processes, extensive studies have been conducted. These studies have demonstrated that a gradient of activities involving multiple signaling pathways specify the embryonic axis and induce cell-type-specific master transcription factors in a spatiotemporal manner. Moreover, applying the knowledge of mesoderm development, researchers have attempted to recapitulate the in vivo development processes in in vitro settings, using mouse and human PSCs. In this review, we summarize the state-of-the-art understanding of mesoderm development and in vitro modeling of mesoderm development using PSCs. We also discuss future perspectives on the use of PSCs to generate skeletal tissues for basic research and clinical applications. -

The Derivatives of Three-Layered Embryo (Germ Layers)
HUMANHUMAN EMBRYOLOGYEMBRYOLOGY Department of Histology and Embryology Jilin University ChapterChapter 22 GeneralGeneral EmbryologyEmbryology FourthFourth week:week: TheThe derivativesderivatives ofof trilaminartrilaminar germgerm discdisc Dorsal side of the germ disc. At the beginning of the third week of development, the ectodermal germ layer has the shape of a disc that is broader in the cephalic than the caudal region. Cross section shows formation of trilaminar germ disc Primitive pit Drawing of a sagittal section through a 17-day embryo. The most cranial portion of the definitive notochord has formed. ectoderm Schematic view showing the definitive notochord. horizon =ectoderm hillside fields =neural plate mountain peaks =neural folds Cave sinks into mountain =neural tube valley =neural groove 7.1 Derivatives of the Ectodermal Germ Layer 1) Formation of neural tube Notochord induces the overlying ectoderm to thicken and form the neural plate. Cross section Animation of formation of neural plate When notochord is forming, primitive streak is shorten. At meanwhile, neural plate is induced to form cephalic to caudal end, following formation of notochord. By the end of 3rd week, neural folds and neural groove are formed. Neural folds fuse in the midline, beginning in cervical region and Cross section proceeding cranially and caudally. Neural tube is formed & invade into the embryo body. A. Dorsal view of a human embryo at approximately day 22. B. Dorsal view of a human embryo at approximately day 23. The nervous system is in connection with the amniotic cavity through the cranial and caudal neuropores. Cranial/anterior neuropore Neural fold heart Neural groove endoderm caudal/posterior neuropore A. -

Nephric Lineage Specification by Pax2 and Pax8
Downloaded from genesdev.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Nephric lineage specification by Pax2 and Pax8 Maxime Bouchard, Abdallah Souabni, Markus Mandler, Annette Neubüser, and Meinrad Busslinger1 Research Institute of Molecular Pathology, Vienna Biocenter, A-1030 Vienna, Austria The mammalian kidney develops in three successive steps from the initial pronephros via the mesonephros to the adult metanephros. Although the nephric lineage is specified during pronephros induction, no single regulator, including the transcription factor Pax2 or Pax8, has yet been identified to control this initial phase of kidney development. In this paper, we demonstrate that mouse embryos lacking both Pax2 and Pax8 are unable to form the pronephros or any later nephric structures. In these double-mutant embryos, the intermediate mesoderm does not undergo the mesenchymal-epithelial transitions required for nephric duct formation, fails to initiate the kidney-specific expression of Lim1and c -Ret, and is lost by apoptosis 1d after failed pronephric induction. Conversely, retroviral misexpression of Pax2 was sufficient to induce ectopic nephric structures in the intermediate mesoderm and genital ridge of chick embryos. Together, these data identify Pax2 and Pax8 as critical regulators that specify the nephric lineage. [Keywords: Pax2;Pax8;genetic redundancy;kidney development;mesenchymal-epithelial transition;lineage specification] Received June 28, 2002;revised version accepted September 20, 2002. Kidney development in mammals and birds proceeds in development (for review, see Kuure et al. 2000;Davies three successive steps that are all characterized by the and Brändli 2002). The majority of these genes are essen- mesenchymal-to-epithelial transformation of intermedi- tial for proper morphogenesis of the metanephros with ate mesoderm cells. -
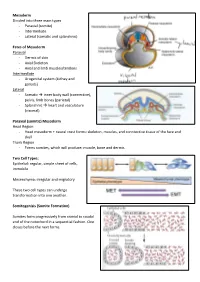
Mesoderm Divided Into Three Main Types - Paraxial (Somite) - Intermediate - Lateral (Somatic and Splanchnic)
Mesoderm Divided into three main types - Paraxial (somite) - Intermediate - Lateral (somatic and splanchnic) Fates of Mesoderm Paraxial - Dermis of skin - Axial Skeleton - Axial and limb muscles/tendons Intermediate - Urogenital system (kidney and gonads) Lateral - Somatic inner body wall (connective), pelvis, limb bones (parietal) - Splanchnic heart and vasculature (visceral) Paraxial (somitic) Mesoderm Head Region - Head mesoderm + neural crest forms: skeleton, muscles, and conntective tissue of the face and skull Trunk Region - Forms somites, which will produce: muscle, bone and dermis Two Cell Types: Epithelial: regular, simple sheet of cells, immobile Mesenchyma: irregular and migratory These two cell types can undergo transformation into one another. Somitogenisis (Somite Formation) Somites form progressively from cranial to caudal end of the notochord in a sequential fashion. One closes before the next forms. Somite Differentiation The somite splits into the epithelial dermamyotome (dermis/muscle) and the messenchymal sclerotome (skeletal). The somite is all paraxial mesoderm. Somite location determines the fate of its associates derma/myo/sclerotomes. Intermediate Mesoderm Urogenital system: - Kidneys - Gonads - Reproductive Duct Systems Runs alongside the paraxial mesoderm. Urogenital System Along mesonephric duct: - Pronephros, mesonephros, and metanephros - Pronephros fall away as gonad develops on ventral-medial side of mesonephros. - Metanephrogenic mesenchyme gives rise to kidney. The mesonephric duct will become the Wolffian duct forming at the nephric bud. The Mullerian duct forms via an invagination on the dorsal side of the nephric duct. The gonad will degenerate one of the two ducts depending on the hormones it produces. XX degenerates Wolffian duct – no testosterone, anti-Mullerian hormone (AMH) not produced, and Mullerian duct can develop in addition to female reproductive organs (ovaries, vagina) XY degenerates Mullerian duct – testosterone, AMH produced, Wolffian duct continues as male reproductive organs (testes, penis) develop. -

Embryology and Teratology in the Curricula of Healthcare Courses
ANATOMICAL EDUCATION Eur. J. Anat. 21 (1): 77-91 (2017) Embryology and Teratology in the Curricula of Healthcare Courses Bernard J. Moxham 1, Hana Brichova 2, Elpida Emmanouil-Nikoloussi 3, Andy R.M. Chirculescu 4 1Cardiff School of Biosciences, Cardiff University, Museum Avenue, Cardiff CF10 3AX, Wales, United Kingdom and Department of Anatomy, St. George’s University, St George, Grenada, 2First Faculty of Medicine, Institute of Histology and Embryology, Charles University Prague, Albertov 4, 128 01 Prague 2, Czech Republic and Second Medical Facul- ty, Institute of Histology and Embryology, Charles University Prague, V Úvalu 84, 150 00 Prague 5 , Czech Republic, 3The School of Medicine, European University Cyprus, 6 Diogenous str, 2404 Engomi, P.O.Box 22006, 1516 Nicosia, Cyprus , 4Department of Morphological Sciences, Division of Anatomy, Faculty of Medicine, C. Davila University, Bucharest, Romania SUMMARY Key words: Anatomy – Embryology – Education – Syllabus – Medical – Dental – Healthcare Significant changes are occurring worldwide in courses for healthcare studies, including medicine INTRODUCTION and dentistry. Critical evaluation of the place, tim- ing, and content of components that can be collec- Embryology is a sub-discipline of developmental tively grouped as the anatomical sciences has biology that relates to life before birth. Teratology however yet to be adequately undertaken. Surveys (τέρατος (teratos) meaning ‘monster’ or ‘marvel’) of teaching hours for embryology in US and UK relates to abnormal development and congenital medical courses clearly demonstrate that a dra- abnormalities (i.e. morphofunctional impairments). matic decline in the importance of the subject is in Embryological studies are concerned essentially progress, in terms of both a decrease in the num- with the laws and mechanisms associated with ber of hours allocated within the medical course normal development (ontogenesis) from the stage and in relation to changes in pedagogic methodol- of the ovum until parturition and the end of intra- ogies. -

Programmed Cell Death in the Developing Somites Is Promoted By
Developmental Biology 228, 326–336 (2000) doi:10.1006/dbio.2000.9948, available online at http://www.idealibrary.com on View metadata, citation and similar papers at core.ac.uk brought to you by CORE Programmed Cell Death in the Developing Somitesprovided by Elsevier - Publisher Connector Is Promoted by Nerve Growth Factor via Its p75NTR Receptor Marı´a L. Cotrina, Maritza Gonza´lez-Hoyuela, Julio A. Barbas, and Alfredo Rodrı´guez-Te´bar1 Instituto Cajal de Neurobiologı´a, CSIC, Avenida Doctor Arce, 37, E-28002 Madrid, Spain Neurotrophins control neuron number during development by promoting the generation and survival of neurons and by regulating programmed neuronal death. In the latter case, the cell death induced by nerve growth factor (NGF) in the developing chick retina is mediated by p75NTR, the common neurotrophin receptor (J. M. Frade, A. Rodriguez-Tebar, and Y.-A. Barde, 1996, Nature 383, 166–168). Here we show that NGF also induces the programmed death of paraxial mesoderm cells in the developing somites. Both NGF and p75NTR are expressed in the somites of chick embryos at the time and the place of programmed cell death. Moreover, neutralizing the activity of endogenous NGF with a specific blocking antibody, or antagonizing NGF binding to p75NTR by the application of human NT-4/5, reduces the levels of apoptotic cell death in both the sclerotome and the dermamyotome by about 50 and 70%, respectively. Previous data have shown that Sonic hedgehog is necessary for the survival of differentiated somite cells. Consistent with this, Sonic hedgehog induces a decrease of NGF mRNA in somite explant cultures, thus showing the antagonistic effect of NGF and Sonic hedgehog with respect to somite cell survival. -

Unit 3 Embryo Questions
Unit 3 Embryology Clinically Oriented Anatomy (COA) Texas Tech University Health Sciences Center Created by Parker McCabe, Fall 2019 parker.mccabe@@uhsc.edu Solu%ons 1. B 11. A 21. D 2. C 12. B 22. D 3. C 13. E 23. D 4. B 14. D 24. A 5. E 15. C 25. D 6. C 16. B 26. B 7. D 17. E 27. C 8. B 18. A 9. C 19. C 10. D 20. B Digestive System 1. Which of the following structures develops as an outgrowth of the endodermal epithelium of the upper part of the duodenum? A. Stomach B. Pancreas C. Lung buds D. Trachea E. Esophagus Ques%on 1 A. Stomach- Foregut endoderm B. Pancreas- The pancreas, liver, and biliary apparatus all develop from outgrowths of the endodermal epithelium of the upper part of the duodenum. C. Lung buds- Foregut endoderm D. Trachea- Foregut endoderm E. Esophagus- Foregut endoderm 2. Where does the spleen originate and then end up after the rotation of abdominal organs during fetal development? A. Ventral mesentery à left side B. Ventral mesentery à right side C. Dorsal mesentery à left side D. Dorsal mesentery à right side E. It does not relocate Question 2 A. Ventral mesentery à left side B. Ventral mesentery à right side C. Dorsal mesentery à left side- The spleen and dorsal pancreas are embedded within the dorsal mesentery (greater omentum). After rotation, dorsal will go to the left side of the body and ventral will go to the right side of the body (except for the ventral pancreas). -

26 April 2010 TE Prepublication Page 1 Nomina Generalia General Terms
26 April 2010 TE PrePublication Page 1 Nomina generalia General terms E1.0.0.0.0.0.1 Modus reproductionis Reproductive mode E1.0.0.0.0.0.2 Reproductio sexualis Sexual reproduction E1.0.0.0.0.0.3 Viviparitas Viviparity E1.0.0.0.0.0.4 Heterogamia Heterogamy E1.0.0.0.0.0.5 Endogamia Endogamy E1.0.0.0.0.0.6 Sequentia reproductionis Reproductive sequence E1.0.0.0.0.0.7 Ovulatio Ovulation E1.0.0.0.0.0.8 Erectio Erection E1.0.0.0.0.0.9 Coitus Coitus; Sexual intercourse E1.0.0.0.0.0.10 Ejaculatio1 Ejaculation E1.0.0.0.0.0.11 Emissio Emission E1.0.0.0.0.0.12 Ejaculatio vera Ejaculation proper E1.0.0.0.0.0.13 Semen Semen; Ejaculate E1.0.0.0.0.0.14 Inseminatio Insemination E1.0.0.0.0.0.15 Fertilisatio Fertilization E1.0.0.0.0.0.16 Fecundatio Fecundation; Impregnation E1.0.0.0.0.0.17 Superfecundatio Superfecundation E1.0.0.0.0.0.18 Superimpregnatio Superimpregnation E1.0.0.0.0.0.19 Superfetatio Superfetation E1.0.0.0.0.0.20 Ontogenesis Ontogeny E1.0.0.0.0.0.21 Ontogenesis praenatalis Prenatal ontogeny E1.0.0.0.0.0.22 Tempus praenatale; Tempus gestationis Prenatal period; Gestation period E1.0.0.0.0.0.23 Vita praenatalis Prenatal life E1.0.0.0.0.0.24 Vita intrauterina Intra-uterine life E1.0.0.0.0.0.25 Embryogenesis2 Embryogenesis; Embryogeny E1.0.0.0.0.0.26 Fetogenesis3 Fetogenesis E1.0.0.0.0.0.27 Tempus natale Birth period E1.0.0.0.0.0.28 Ontogenesis postnatalis Postnatal ontogeny E1.0.0.0.0.0.29 Vita postnatalis Postnatal life E1.0.1.0.0.0.1 Mensurae embryonicae et fetales4 Embryonic and fetal measurements E1.0.1.0.0.0.2 Aetas a fecundatione5 Fertilization -

A Coordinated Progression of Progenitor Cell States Initiates Urinary Tract Development
ARTICLE https://doi.org/10.1038/s41467-021-22931-5 OPEN A coordinated progression of progenitor cell states initiates urinary tract development Oraly Sanchez-Ferras 1, Alain Pacis1,2, Maria Sotiropoulou3, Yuhong Zhang1, Yu Chang Wang3, ✉ Mathieu Bourgey2,3, Guillaume Bourque 2,3, Jiannis Ragoussis 3,4 & Maxime Bouchard 1 The kidney and upper urinary tract develop through reciprocal interactions between the ureteric bud and the surrounding mesenchyme. Ureteric bud branching forms the arborized 1234567890():,; collecting duct system of the kidney, while ureteric tips promote nephron formation from dedicated progenitor cells. While nephron progenitor cells are relatively well characterized, the origin of ureteric bud progenitors has received little attention so far. It is well established that the ureteric bud is induced from the nephric duct, an epithelial duct derived from the intermediate mesoderm of the embryo. However, the cell state transitions underlying the progression from intermediate mesoderm to nephric duct and ureteric bud remain unknown. Here we show that nephric duct morphogenesis results from the coordinated organization of four major progenitor cell populations. Using single cell RNA-seq and Cluster RNA-seq, we show that these progenitors emerge in time and space according to a stereotypical pattern. We identify the transcription factors Tfap2a/b and Gata3 as critical coordinators of this progenitor cell progression. This study provides a better understanding of the cellular origin of the renal collecting duct system and associated urinary tract developmental diseases, which may inform guided differentiation of functional kidney tissue. 1 Goodman Cancer Research Centre and Department of Biochemistry, McGill University, Montreal, QC, Canada. 2 Canadian Centre for Computational Genomics, McGill University, Montréal, QC, Canada. -

COUP-TFII Is Essential for Metanephric Mesenchyme Formation and Kidney Precursor Cell Survival Cheng-Tai Yu1, Ke Tang1, Jae Mi Suh1,*, Rulang Jiang3, Sophia Y
2330 RESEARCH ARTICLE Development 139, 2330-2339 (2012) doi:10.1242/dev.076299 © 2012. Published by The Company of Biologists Ltd COUP-TFII is essential for metanephric mesenchyme formation and kidney precursor cell survival Cheng-Tai Yu1, Ke Tang1, Jae Mi Suh1,*, Rulang Jiang3, Sophia Y. Tsai1,2,‡ and Ming-Jer Tsai1,2,‡ SUMMARY Development of the metanephric kidney in mammals requires complex reciprocal tissue interactions between the ureteric epithelium and the mesenchyme. It is believed that Gdnf, produced in the metanephric mesenchyme, activates Ret signaling in the Wolffian duct to initiate the formation of the metanephros. However, the molecular mechanism for induction of Gdnf in the metanephric mesenchyme is not completely defined. Previous studies demonstrated that during the early stages of kidney development, loss of Osr1, Eya1, Pax2 or Wt1 gene function in the metanephric mesenchyme compromises the formation of the kidney. Moreover, it has been shown that the Hox11-Eya1-Pax2 complex activates the expression of Six2 and Gdnf in the metanephric mesenchyme to drive nephrogenesis. Here, we demonstrate that the orphan nuclear receptor chicken ovalbumin upstream promoter transcription factor II (COUP-TFII, also known as Nr2f2) is required for the specification of the metanephric mesenchyme. Deletion of COUP-TFII at E7.5 results in improper differentiation of the metanephric mesenchyme and absence of essential developmental regulators, such as Eya1, Six2, Pax2 and Gdnf. Importantly, we show that COUP-TFII directly regulates the expression of both Eya1 and Wt1 in the metanephric mesenchyme. Our findings reveal, for the first time, that COUP-TFII plays a central role in the specification of metanephric fate and in the maintenance of metanephric mesenchyme proliferation and survival by acting as a crucial regulator of Eya1 and Wt1 expression. -

Urogenital Development
2-5-03 Urogenital Development Greg Dressler Assoc. Professor Dept. of Pathology x46490 [email protected] The Origin of the Kidney In the vertebrate embryo, the first stage of kidney development occurs after gastrulation, within a region called the intermediate mesoderm. The process of gastrulation turns a single sheet of pluripotent embryonic Ectoderm, or the epiblast, into three primary germ layers. 1 The Origin of the Kidney In the vertebrate embryo, the first stage of kidney development occurs after gastrulation, within a region called the intermediate mesoderm. The process of gastrulation turns a single sheet of pluripotent embryonic Ectoderm, or the epiblast, into three primary germ layers. Neural tube Neural crest Muscle, Bone, Skin Ectoderm Paraxial Mesoderm Kidney, Gonads, Epiblast Mesoderm Intermediate Mesoderm Reproductive organs Lateral plate Mesoderm Mesenteries, Hemangioblastic Endoderm Heart, limbs, etc. Gut, pancreas, liver, lung, etc. Cross sections through progressive stages of the chick reveal the organization of the early mesoderm The primitive streak is a furrow through which epiblast cells migrate The distance from the neural plate, the embryonic axis or midline, determines the type of mesoderm and its eventual fate The midline NOTOCHORD secretes factors that organize the dorsal-ventral and medio-lateral axes of the embryo. 2 The first epithelial component of the urogenital system is the NEPHRIC DUCT. It arises within the intermediate mesoderm adjacent to the 10-12th somites and extends posteriorly. DiI lineage tracing in the chick embryo reveals that the Nephric Duct forms by extension rather than by recruitment of mesoderm to the epithelial duct. Obara-Ishihara et al., Develop.