Directing Human Embryonic Stem Cell Differentiation Towards a Renal Lineage Generates a Self-Organizing Kidney
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

3 Embryology and Development
BIOL 6505 − INTRODUCTION TO FETAL MEDICINE 3. EMBRYOLOGY AND DEVELOPMENT Arlet G. Kurkchubasche, M.D. INTRODUCTION Embryology – the field of study that pertains to the developing organism/human Basic embryology –usually taught in the chronologic sequence of events. These events are the basis for understanding the congenital anomalies that we encounter in the fetus, and help explain the relationships to other organ system concerns. Below is a synopsis of some of the critical steps in embryogenesis from the anatomic rather than molecular basis. These concepts will be more intuitive and evident in conjunction with diagrams and animated sequences. This text is a synopsis of material provided in Langman’s Medical Embryology, 9th ed. First week – ovulation to fertilization to implantation Fertilization restores 1) the diploid number of chromosomes, 2) determines the chromosomal sex and 3) initiates cleavage. Cleavage of the fertilized ovum results in mitotic divisions generating blastomeres that form a 16-cell morula. The dense morula develops a central cavity and now forms the blastocyst, which restructures into 2 components. The inner cell mass forms the embryoblast and outer cell mass the trophoblast. Consequences for fetal management: Variances in cleavage, i.e. splitting of the zygote at various stages/locations - leads to monozygotic twinning with various relationships of the fetal membranes. Cleavage at later weeks will lead to conjoined twinning. Second week: the week of twos – marked by bilaminar germ disc formation. Commences with blastocyst partially embedded in endometrial stroma Trophoblast forms – 1) cytotrophoblast – mitotic cells that coalesce to form 2) syncytiotrophoblast – erodes into maternal tissues, forms lacunae which are critical to development of the uteroplacental circulation. -

The Rotated Hypoblast of the Chicken Embryo Does Not Initiate an Ectopic Axis in the Epiblast
Proc. Natl. Acad. Sci. USA Vol. 92, pp. 10733-10737, November 1995 Developmental Biology The rotated hypoblast of the chicken embryo does not initiate an ectopic axis in the epiblast ODED KHANER Department of Cell and Animal Biology, The Hebrew University, Jerusalem, Israel 91904 Communicated by John Gerhart, University of California, Berkeley, CA, July 14, 1995 ABSTRACT In the amniotes, two unique layers of cells, of the hypoblast and that the orientation of the streak and axis the epiblast and the hypoblast, constitute the embryo at the can be predicted from the polarity of the stage XIII epiblast. blastula stage. All the tissues of the adult will derive from the Still it was thought that the polarity of the epiblast is sufficient epiblast, whereas hypoblast cells will form extraembryonic for axial development in experimental situations, although in yolk sac endoderm. During gastrulation, the endoderm and normal development the polarity of the hypoblast is dominant the mesoderm of the embryo arise from the primitive streak, (5, 6). These reports (1-3, 5, 6), taken together, would imply which is an epiblast structure through which cells enter the that cells of the hypoblast not only have the ability to change interior. Previous investigations by others have led to the the fate of competent cells in the epiblast to initiate an ectopic conclusion that the avian hypoblast, when rotated with regard axis, but also have the ability to repress the formation of the to the epiblast, has inductive properties that can change the original axis in committed cells of the epiblast. At the same fate of competent cells in the epiblast to form an ectopic time, the results showed that the epiblast is not wholly depen- embryonic axis. -

Gastrulation
Embryology of the spine and spinal cord Andrea Rossi, MD Neuroradiology Unit Istituto Giannina Gaslini Hospital Genoa, Italy [email protected] LEARNING OBJECTIVES: LEARNING OBJECTIVES: 1) To understand the basics of spinal 1) To understand the basics of spinal cord development cord development 2) To understand the general rules of the 2) To understand the general rules of the development of the spine development of the spine 3) To understand the peculiar variations 3) To understand the peculiar variations to the normal spine plan that occur at to the normal spine plan that occur at the CVJ the CVJ Summary of week 1 Week 2-3 GASTRULATION "It is not birth, marriage, or death, but gastrulation, which is truly the most important time in your life." Lewis Wolpert (1986) Gastrulation Conversion of the embryonic disk from a bilaminar to a trilaminar arrangement and establishment of the notochord The three primary germ layers are established The basic body plan is established, including the physical construction of the rudimentary primary body axes As a result of the movements of gastrulation, cells are brought into new positions, allowing them to interact with cells that were initially not near them. This paves the way for inductive interactions, which are the hallmark of neurulation and organogenesis Day 16 H E Day 15 Dorsal view of a 0.4 mm embryo BILAMINAR DISK CRANIAL Epiblast faces the amniotic sac node Hypoblast Primitive pit (primitive endoderm) faces the yolk sac Primitive streak CAUDAL Prospective notochordal cells Dias Dias During -

The Genetic Basis of Mammalian Neurulation
REVIEWS THE GENETIC BASIS OF MAMMALIAN NEURULATION Andrew J. Copp*, Nicholas D. E. Greene* and Jennifer N. Murdoch‡ More than 80 mutant mouse genes disrupt neurulation and allow an in-depth analysis of the underlying developmental mechanisms. Although many of the genetic mutants have been studied in only rudimentary detail, several molecular pathways can already be identified as crucial for normal neurulation. These include the planar cell-polarity pathway, which is required for the initiation of neural tube closure, and the sonic hedgehog signalling pathway that regulates neural plate bending. Mutant mice also offer an opportunity to unravel the mechanisms by which folic acid prevents neural tube defects, and to develop new therapies for folate-resistant defects. 6 ECTODERM Neurulation is a fundamental event of embryogenesis distinct locations in the brain and spinal cord .By The outer of the three that culminates in the formation of the neural tube, contrast, the mechanisms that underlie the forma- embryonic (germ) layers that which is the precursor of the brain and spinal cord. A tion, elevation and fusion of the neural folds have gives rise to the entire central region of specialized dorsal ECTODERM, the neural plate, remained elusive. nervous system, plus other organs and embryonic develops bilateral neural folds at its junction with sur- An opportunity has now arisen for an incisive analy- structures. face (non-neural) ectoderm. These folds elevate, come sis of neurulation mechanisms using the growing battery into contact (appose) in the midline and fuse to create of genetically targeted and other mutant mouse strains NEURAL CREST the neural tube, which, thereafter, becomes covered by in which NTDs form part of the mutant phenotype7.At A migratory cell population that future epidermal ectoderm. -

Understanding Paraxial Mesoderm Development and Sclerotome Specification for Skeletal Repair Shoichiro Tani 1,2, Ung-Il Chung2,3, Shinsuke Ohba4 and Hironori Hojo2,3
Tani et al. Experimental & Molecular Medicine (2020) 52:1166–1177 https://doi.org/10.1038/s12276-020-0482-1 Experimental & Molecular Medicine REVIEW ARTICLE Open Access Understanding paraxial mesoderm development and sclerotome specification for skeletal repair Shoichiro Tani 1,2, Ung-il Chung2,3, Shinsuke Ohba4 and Hironori Hojo2,3 Abstract Pluripotent stem cells (PSCs) are attractive regenerative therapy tools for skeletal tissues. However, a deep understanding of skeletal development is required in order to model this development with PSCs, and for the application of PSCs in clinical settings. Skeletal tissues originate from three types of cell populations: the paraxial mesoderm, lateral plate mesoderm, and neural crest. The paraxial mesoderm gives rise to the sclerotome mainly through somitogenesis. In this process, key developmental processes, including initiation of the segmentation clock, formation of the determination front, and the mesenchymal–epithelial transition, are sequentially coordinated. The sclerotome further forms vertebral columns and contributes to various other tissues, such as tendons, vessels (including the dorsal aorta), and even meninges. To understand the molecular mechanisms underlying these developmental processes, extensive studies have been conducted. These studies have demonstrated that a gradient of activities involving multiple signaling pathways specify the embryonic axis and induce cell-type-specific master transcription factors in a spatiotemporal manner. Moreover, applying the knowledge of mesoderm development, researchers have attempted to recapitulate the in vivo development processes in in vitro settings, using mouse and human PSCs. In this review, we summarize the state-of-the-art understanding of mesoderm development and in vitro modeling of mesoderm development using PSCs. We also discuss future perspectives on the use of PSCs to generate skeletal tissues for basic research and clinical applications. -

Sonic Hedgehog a Neural Tube Anti-Apoptotic Factor 4013 Other Side of the Neural Plate, Remaining in Contact with Midline Cells, RESULTS Was Used As a Control
Development 128, 4011-4020 (2001) 4011 Printed in Great Britain © The Company of Biologists Limited 2001 DEV2740 Anti-apoptotic role of Sonic hedgehog protein at the early stages of nervous system organogenesis Jean-Baptiste Charrier, Françoise Lapointe, Nicole M. Le Douarin and Marie-Aimée Teillet* Institut d’Embryologie Cellulaire et Moléculaire, CNRS FRE2160, 49bis Avenue de la Belle Gabrielle, 94736 Nogent-sur-Marne Cedex, France *Author for correspondence (e-mail: [email protected]) Accepted 19 July 2001 SUMMARY In vertebrates the neural tube, like most of the embryonic notochord or a floor plate fragment in its vicinity. The organs, shows discreet areas of programmed cell death at neural tube can also be recovered by transplanting it into several stages during development. In the chick embryo, a stage-matched chick embryo having one of these cell death is dramatically increased in the developing structures. In addition, cells engineered to produce Sonic nervous system and other tissues when the midline cells, hedgehog protein (SHH) can mimic the effect of the notochord and floor plate, are prevented from forming by notochord and floor plate cells in in situ grafts and excision of the axial-paraxial hinge (APH), i.e. caudal transplantation experiments. SHH can thus counteract a Hensen’s node and rostral primitive streak, at the 6-somite built-in cell death program and thereby contribute to organ stage (Charrier, J. B., Teillet, M.-A., Lapointe, F. and Le morphogenesis, in particular in the central nervous system. Douarin, N. M. (1999). Development 126, 4771-4783). In this paper we demonstrate that one day after APH excision, Key words: Apoptosis, Avian embryo, Cell death, Cell survival, when dramatic apoptosis is already present in the neural Floor plate, Notochord, Quail/chick, Shh, Somite, Neural tube, tube, the latter can be rescued from death by grafting a Spinal cord INTRODUCTION generally induces an inflammatory response. -

The Derivatives of Three-Layered Embryo (Germ Layers)
HUMANHUMAN EMBRYOLOGYEMBRYOLOGY Department of Histology and Embryology Jilin University ChapterChapter 22 GeneralGeneral EmbryologyEmbryology FourthFourth week:week: TheThe derivativesderivatives ofof trilaminartrilaminar germgerm discdisc Dorsal side of the germ disc. At the beginning of the third week of development, the ectodermal germ layer has the shape of a disc that is broader in the cephalic than the caudal region. Cross section shows formation of trilaminar germ disc Primitive pit Drawing of a sagittal section through a 17-day embryo. The most cranial portion of the definitive notochord has formed. ectoderm Schematic view showing the definitive notochord. horizon =ectoderm hillside fields =neural plate mountain peaks =neural folds Cave sinks into mountain =neural tube valley =neural groove 7.1 Derivatives of the Ectodermal Germ Layer 1) Formation of neural tube Notochord induces the overlying ectoderm to thicken and form the neural plate. Cross section Animation of formation of neural plate When notochord is forming, primitive streak is shorten. At meanwhile, neural plate is induced to form cephalic to caudal end, following formation of notochord. By the end of 3rd week, neural folds and neural groove are formed. Neural folds fuse in the midline, beginning in cervical region and Cross section proceeding cranially and caudally. Neural tube is formed & invade into the embryo body. A. Dorsal view of a human embryo at approximately day 22. B. Dorsal view of a human embryo at approximately day 23. The nervous system is in connection with the amniotic cavity through the cranial and caudal neuropores. Cranial/anterior neuropore Neural fold heart Neural groove endoderm caudal/posterior neuropore A. -

The Origin of Early Primitive Streak 89
Development 127, 87-96 (2000) 87 Printed in Great Britain © The Company of Biologists Limited 2000 DEV3080 Formation of the avian primitive streak from spatially restricted blastoderm: evidence for polarized cell division in the elongating streak Yan Wei and Takashi Mikawa* Department of Cell Biology, Cornell University Medical College, 1300 York Avenue, New York, NY 10021, USA *Author for correspondence (e-mail: [email protected]) Accepted 13 October; published on WWW 8 December 1999 SUMMARY Gastrulation in the amniote begins with the formation of a cells generated daughter cells that underwent a polarized primitive streak through which precursors of definitive cell division oriented perpendicular to the anteroposterior mesoderm and endoderm ingress and migrate to their embryonic axis. The resulting daughter cell population was embryonic destinations. This organizing center for amniote arranged in a longitudinal array extending the complete gastrulation is induced by signal(s) from the posterior length of the primitive streak. Furthermore, expression of margin of the blastodisc. The mode of action of these cVg1, a posterior margin-derived signal, at the anterior inductive signal(s) remains unresolved, since various marginal zone induced adjacent epiblast cells, but not those origins and developmental pathways of the primitive streak lateral to or distant from the signal, to form an ectopic have been proposed. In the present study, the fate of primitive streak. The cVg1-induced epiblast cells also chicken blastodermal cells was traced for the first time in exhibited polarized cell divisions during ectopic primitive ovo from prestreak stages XI-XII through HH stage 3, streak formation. These results suggest that blastoderm when the primitive streak is initially established and prior cells located immediately anterior to the posterior marginal to the migration of mesoderm. -

Nephric Lineage Specification by Pax2 and Pax8
Downloaded from genesdev.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Nephric lineage specification by Pax2 and Pax8 Maxime Bouchard, Abdallah Souabni, Markus Mandler, Annette Neubüser, and Meinrad Busslinger1 Research Institute of Molecular Pathology, Vienna Biocenter, A-1030 Vienna, Austria The mammalian kidney develops in three successive steps from the initial pronephros via the mesonephros to the adult metanephros. Although the nephric lineage is specified during pronephros induction, no single regulator, including the transcription factor Pax2 or Pax8, has yet been identified to control this initial phase of kidney development. In this paper, we demonstrate that mouse embryos lacking both Pax2 and Pax8 are unable to form the pronephros or any later nephric structures. In these double-mutant embryos, the intermediate mesoderm does not undergo the mesenchymal-epithelial transitions required for nephric duct formation, fails to initiate the kidney-specific expression of Lim1and c -Ret, and is lost by apoptosis 1d after failed pronephric induction. Conversely, retroviral misexpression of Pax2 was sufficient to induce ectopic nephric structures in the intermediate mesoderm and genital ridge of chick embryos. Together, these data identify Pax2 and Pax8 as critical regulators that specify the nephric lineage. [Keywords: Pax2;Pax8;genetic redundancy;kidney development;mesenchymal-epithelial transition;lineage specification] Received June 28, 2002;revised version accepted September 20, 2002. Kidney development in mammals and birds proceeds in development (for review, see Kuure et al. 2000;Davies three successive steps that are all characterized by the and Brändli 2002). The majority of these genes are essen- mesenchymal-to-epithelial transformation of intermedi- tial for proper morphogenesis of the metanephros with ate mesoderm cells. -
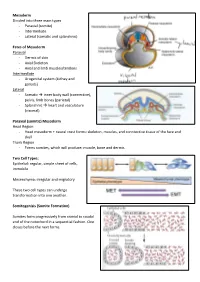
Mesoderm Divided Into Three Main Types - Paraxial (Somite) - Intermediate - Lateral (Somatic and Splanchnic)
Mesoderm Divided into three main types - Paraxial (somite) - Intermediate - Lateral (somatic and splanchnic) Fates of Mesoderm Paraxial - Dermis of skin - Axial Skeleton - Axial and limb muscles/tendons Intermediate - Urogenital system (kidney and gonads) Lateral - Somatic inner body wall (connective), pelvis, limb bones (parietal) - Splanchnic heart and vasculature (visceral) Paraxial (somitic) Mesoderm Head Region - Head mesoderm + neural crest forms: skeleton, muscles, and conntective tissue of the face and skull Trunk Region - Forms somites, which will produce: muscle, bone and dermis Two Cell Types: Epithelial: regular, simple sheet of cells, immobile Mesenchyma: irregular and migratory These two cell types can undergo transformation into one another. Somitogenisis (Somite Formation) Somites form progressively from cranial to caudal end of the notochord in a sequential fashion. One closes before the next forms. Somite Differentiation The somite splits into the epithelial dermamyotome (dermis/muscle) and the messenchymal sclerotome (skeletal). The somite is all paraxial mesoderm. Somite location determines the fate of its associates derma/myo/sclerotomes. Intermediate Mesoderm Urogenital system: - Kidneys - Gonads - Reproductive Duct Systems Runs alongside the paraxial mesoderm. Urogenital System Along mesonephric duct: - Pronephros, mesonephros, and metanephros - Pronephros fall away as gonad develops on ventral-medial side of mesonephros. - Metanephrogenic mesenchyme gives rise to kidney. The mesonephric duct will become the Wolffian duct forming at the nephric bud. The Mullerian duct forms via an invagination on the dorsal side of the nephric duct. The gonad will degenerate one of the two ducts depending on the hormones it produces. XX degenerates Wolffian duct – no testosterone, anti-Mullerian hormone (AMH) not produced, and Mullerian duct can develop in addition to female reproductive organs (ovaries, vagina) XY degenerates Mullerian duct – testosterone, AMH produced, Wolffian duct continues as male reproductive organs (testes, penis) develop. -

A Review of Primitive Streak Formation and Somitogenesis
SPATIOTEMPORAL PATTERN FORMATION IN EARLY DEVELOPMENT: A REVIEW OF PRIMITIVE STREAK FORMATION AND SOMITOGENESIS S. SCHNELL', K.J. PAINTERt, P.K. MAINI' , AND H.G. OTHMERt Abstract. The basic body plan of a number of vertebrates results from two pro cesses that occur early in the development of the blastoderm: large scale rearrangements of tissue via a process called gastrulation, and axial subdivision of tissue in a process called somitogenesis. The first step of gastrulation in avians is formation of the prim itive streak, which marks the first clear manifestation of the anterior-posterior axis. Cell movements that occur through the streak ultimately convert the single layered blastoderm into a trilaminar blastoderm comprising prospective endodermal, mesoder mal and ectodermal tissue. During streak formation a group of cells moves anteriorly as a coherent column from the posterior end of the blastoderm, and as it proceeds other cells stream over the lateral edges of the furrow left behind. The anterior end of the streak is a specialized structure called Hensen's node, which serves as an organizing center for later axis formation and determination of the left-right asymmetry of the body. Soon after the primitive streak forms, Hensen's node regresses towards the tail, leaving the notochord and a pair of segmental plates parallel to the primitive streak in its wake. The posterior end of the segmental plate moves down the cranio-caudal axis with the node, as more cells are added to it by cell division within the plate and by cells entering from the primitive streak. A pair of somites forms from the anterior ends of the two plates at regular intervals. -

The Hypoblast (Visceral Endoderm): an Evo-Devo Perspective Claudio D
REVIEW 1059 Development 139, 1059-1069 (2012) doi:10.1242/dev.070730 © 2012. Published by The Company of Biologists Ltd The hypoblast (visceral endoderm): an evo-devo perspective Claudio D. Stern1,* and Karen M. Downs2 Summary animals develop very quickly; their early cleavages occur without When amniotes appeared during evolution, embryos freed G1 or G2 phases and consist only of mitotic (M) and DNA synthetic themselves from intracellular nutrition; development slowed, (S) phases, and they rely upon maternal mRNAs and proteins until the mid-blastula transition was lost and maternal components zygotic gene expression is activated, usually at around the 11th cell became less important for polarity. Extra-embryonic tissues division (2024 cells). Rapid development and absence of growth emerged to provide nutrition and other innovations. One such imply that the embryonic axes or body plan must be set up quite tissue, the hypoblast (visceral endoderm in mouse), acquired a quickly, within about 24 hours of fertilization, and by subdivision of role in fixing the body plan: it controls epiblast cell movements the original mass of protoplasm, as seen in long germ band insects, leading to primitive streak formation, generating bilateral such as Drosophila. Because of the absence of early growth, the symmetry. It also transiently induces expression of pre-neural fertilized egg of anamniotes contains all the nutrients necessary to markers in the epiblast, which also contributes to delay streak sustain the embryo until it can feed itself, some time after hatching. formation. After gastrulation, the hypoblast might protect In amphibians, nutrition is provided by intracellular yolk, and in prospective forebrain cells from caudalizing signals.