A Coordinated Progression of Progenitor Cell States Initiates Urinary Tract Development
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

3 Embryology and Development
BIOL 6505 − INTRODUCTION TO FETAL MEDICINE 3. EMBRYOLOGY AND DEVELOPMENT Arlet G. Kurkchubasche, M.D. INTRODUCTION Embryology – the field of study that pertains to the developing organism/human Basic embryology –usually taught in the chronologic sequence of events. These events are the basis for understanding the congenital anomalies that we encounter in the fetus, and help explain the relationships to other organ system concerns. Below is a synopsis of some of the critical steps in embryogenesis from the anatomic rather than molecular basis. These concepts will be more intuitive and evident in conjunction with diagrams and animated sequences. This text is a synopsis of material provided in Langman’s Medical Embryology, 9th ed. First week – ovulation to fertilization to implantation Fertilization restores 1) the diploid number of chromosomes, 2) determines the chromosomal sex and 3) initiates cleavage. Cleavage of the fertilized ovum results in mitotic divisions generating blastomeres that form a 16-cell morula. The dense morula develops a central cavity and now forms the blastocyst, which restructures into 2 components. The inner cell mass forms the embryoblast and outer cell mass the trophoblast. Consequences for fetal management: Variances in cleavage, i.e. splitting of the zygote at various stages/locations - leads to monozygotic twinning with various relationships of the fetal membranes. Cleavage at later weeks will lead to conjoined twinning. Second week: the week of twos – marked by bilaminar germ disc formation. Commences with blastocyst partially embedded in endometrial stroma Trophoblast forms – 1) cytotrophoblast – mitotic cells that coalesce to form 2) syncytiotrophoblast – erodes into maternal tissues, forms lacunae which are critical to development of the uteroplacental circulation. -

ROBO2 Restricts the Nephrogenic Field and Regulates Wolffian Duct–Nephrogenic Cord Separation
Developmental Biology 404 (2015) 88–102 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/locate/developmentalbiology ROBO2 restricts the nephrogenic field and regulates Wolffian duct–nephrogenic cord separation Elanor N. Wainwright 1, Dagmar Wilhelm 2, Alexander N. Combes 3, Melissa H. Little 4, Peter Koopman n Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD 4072, Australia article info abstract Article history: ROBO2 plays a key role in regulating ureteric bud (UB) formation in the embryo, with mutations in Received 7 April 2015 humans and mice leading to supernumerary kidneys. Previous studies have established that the number Received in revised form and position of UB outgrowths is determined by the domain of metanephric mesenchymal Gdnf ex- 28 May 2015 pression, which is expanded anteriorly in Robo2 mouse mutants. To clarify how this phenotype arises, we Accepted 30 May 2015 used high-resolution 3D imaging to reveal an increase in the number of nephrogenic cord cells, leading Available online 23 June 2015 to extension of the metanephric mesenchyme field in Robo2-null mouse embryos. Ex vivo experiments Keywords: suggested a dependence of this effect on proliferative signals from the Wolffian duct. Loss of Robo2 Kidney resulted in a failure of the normal separation of the mesenchyme from the Wolffian duct/ureteric epi- Wolffian duct thelium, suggesting that aberrant juxtaposition of these two compartments in Robo2-null mice exposes Ureteric bud the mesenchyme to abnormally high levels of proliferative stimuli. Our data suggest a new model in Nephrogenic cord Mouse which SLIT-ROBO signalling acts not by attenuating Gdnf expression or activity, but instead by limiting epithelial/mesenchymal interactions in the nascent metanephros and restricting the extent of the ne- phrogenic field. -

Understanding Paraxial Mesoderm Development and Sclerotome Specification for Skeletal Repair Shoichiro Tani 1,2, Ung-Il Chung2,3, Shinsuke Ohba4 and Hironori Hojo2,3
Tani et al. Experimental & Molecular Medicine (2020) 52:1166–1177 https://doi.org/10.1038/s12276-020-0482-1 Experimental & Molecular Medicine REVIEW ARTICLE Open Access Understanding paraxial mesoderm development and sclerotome specification for skeletal repair Shoichiro Tani 1,2, Ung-il Chung2,3, Shinsuke Ohba4 and Hironori Hojo2,3 Abstract Pluripotent stem cells (PSCs) are attractive regenerative therapy tools for skeletal tissues. However, a deep understanding of skeletal development is required in order to model this development with PSCs, and for the application of PSCs in clinical settings. Skeletal tissues originate from three types of cell populations: the paraxial mesoderm, lateral plate mesoderm, and neural crest. The paraxial mesoderm gives rise to the sclerotome mainly through somitogenesis. In this process, key developmental processes, including initiation of the segmentation clock, formation of the determination front, and the mesenchymal–epithelial transition, are sequentially coordinated. The sclerotome further forms vertebral columns and contributes to various other tissues, such as tendons, vessels (including the dorsal aorta), and even meninges. To understand the molecular mechanisms underlying these developmental processes, extensive studies have been conducted. These studies have demonstrated that a gradient of activities involving multiple signaling pathways specify the embryonic axis and induce cell-type-specific master transcription factors in a spatiotemporal manner. Moreover, applying the knowledge of mesoderm development, researchers have attempted to recapitulate the in vivo development processes in in vitro settings, using mouse and human PSCs. In this review, we summarize the state-of-the-art understanding of mesoderm development and in vitro modeling of mesoderm development using PSCs. We also discuss future perspectives on the use of PSCs to generate skeletal tissues for basic research and clinical applications. -

Urinary System Intermediate Mesoderm
Urinary System Intermediate mesoderm lateral mesoderm: somite ectoderm neural NOTE: Intermediate mesoderm splanchnic groove somatic is situated between somites and lateral mesoderm (somatic and splanchnic mesoderm bordering the coelom). All mesoderm is derived from the primary mesen- intermediate mesoderm endoderm chyme that migrated through the notochord coelom (becomes urogenital ridge) primitive streak. Intermediate mesoderm (plus adjacent mesothelium lining the coelom) forms a urogenital ridge, which consists of a laterally-positioned nephrogenic cord (that forms kidneys & ureter) and a medially-positioned gonadal ridge (for ovary/testis & female/male genital tract formation). Thus urinary & genital systems have a common embryonic origin; also, they share common ducts. NOTE: Urine production essentially requires an increased capillary surface area (glomeruli), epithelial tubules to collect plasma filtrate and extract desirable constituents, and a duct system to convey urine away from the body. Kidneys Bilateraly, three kid- mesonephric duct neys develop from the neph- metanephros pronephros rogenic cord. They develop mesonephric tubules chronologically in cranial- mesonephros caudal sequence, and are designated pro—, meso—, Nephrogenic Cord (left) and meta—, respectively. cloaca The pronephros and mesonephros develop similarly: the nephrogenic cord undergoes seg- mentation, segments become tubules, tubules drain into a duct & eventually tubules disintegrate. spinal ganglion 1] Pronephros—consists of (7-8) primitive tubules and a pronephric duct that grows caudally and terminates in the cloaca. The tubules soon degenerate, but the pronephric duct persists as the neural tube mesonephric duct. (The pronephros is not functional, somite except in sheep.) notochord mesonephric NOTE tubule The mesonephros is the functional kidney for fish and am- aorta phibians. The metanephros is the functional kidney body of reptiles, birds, & mammals. -

The Derivatives of Three-Layered Embryo (Germ Layers)
HUMANHUMAN EMBRYOLOGYEMBRYOLOGY Department of Histology and Embryology Jilin University ChapterChapter 22 GeneralGeneral EmbryologyEmbryology FourthFourth week:week: TheThe derivativesderivatives ofof trilaminartrilaminar germgerm discdisc Dorsal side of the germ disc. At the beginning of the third week of development, the ectodermal germ layer has the shape of a disc that is broader in the cephalic than the caudal region. Cross section shows formation of trilaminar germ disc Primitive pit Drawing of a sagittal section through a 17-day embryo. The most cranial portion of the definitive notochord has formed. ectoderm Schematic view showing the definitive notochord. horizon =ectoderm hillside fields =neural plate mountain peaks =neural folds Cave sinks into mountain =neural tube valley =neural groove 7.1 Derivatives of the Ectodermal Germ Layer 1) Formation of neural tube Notochord induces the overlying ectoderm to thicken and form the neural plate. Cross section Animation of formation of neural plate When notochord is forming, primitive streak is shorten. At meanwhile, neural plate is induced to form cephalic to caudal end, following formation of notochord. By the end of 3rd week, neural folds and neural groove are formed. Neural folds fuse in the midline, beginning in cervical region and Cross section proceeding cranially and caudally. Neural tube is formed & invade into the embryo body. A. Dorsal view of a human embryo at approximately day 22. B. Dorsal view of a human embryo at approximately day 23. The nervous system is in connection with the amniotic cavity through the cranial and caudal neuropores. Cranial/anterior neuropore Neural fold heart Neural groove endoderm caudal/posterior neuropore A. -

Embryology of the Kidney Rizaldy Paz Scott | Yoshiro Maezawa | Jordan Kreidberg | Susan E
1 Embryology of the Kidney Rizaldy Paz Scott | Yoshiro Maezawa | Jordan Kreidberg | Susan E. Quaggin CHAPTER OUTLINE MAMMALIAN KIDNEY DEVELOPMENT, 2 MOLECULAR GENETICS OF MODEL SYSTEMS TO STUDY KIDNEY NEPHROGENESIS, 22 DEVELOPMENT, 8 GENETIC ANALYSIS OF MAMMALIAN KIDNEY DEVELOPMENT, 15 KEY POINTS • The development of the kidney relies on reciprocal signaling and inductive interactions between neighboring cells. • Epithelial cells that comprise the tubular structures of the kidney are derived from two distinct cell lineages: the ureteric epithelia lineage that branches and gives rise to collecting ducts and the nephrogenic mesenchyme lineage that undergoes mesenchyme to epithelial transition to form connecting tubules, distal tubules, the loop of Henle, proximal tubules, parietal epithelial cells, and podocytes. • Nephrogenesis and nephron endowment requires an epigenetically regulated balance between nephron progenitor self-renewal and epithelial differentiation. • The timing of incorporation of nephron progenitor cells into nascent nephrons predicts their positional identity within the highly patterned mature nephron. • Stromal cells and their derivatives coregulate ureteric branching morphogenesis, nephrogenesis, and vascular development. • Endothelial cells track the development of the ureteric epithelia and establish the renal vasculature through a combination of vasculogenic and angiogenic processes. • Collecting duct epithelia have an inherent plasticity enabling them to switch between principal and intercalated cell identities. MAMMALIAN KIDNEY DEVELOPMENT The filtration function of the kidneys is accomplished by basic units called nephrons (Fig. 1.1). Humans on average have 1 million nephrons per adult kidney but the range of ANATOMIC OVERVIEW OF THE 4 MAMMALIAN KIDNEY total nephrons is highly variable across human populations. Each mouse kidney may contain up to 12,000–16,000 nephrons The kidney is a sophisticated, highly vascularized organ that depending on the strain.5 This wide range in nephron number plays a central role in overall body homeostasis. -

Nephric Lineage Specification by Pax2 and Pax8
Downloaded from genesdev.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Nephric lineage specification by Pax2 and Pax8 Maxime Bouchard, Abdallah Souabni, Markus Mandler, Annette Neubüser, and Meinrad Busslinger1 Research Institute of Molecular Pathology, Vienna Biocenter, A-1030 Vienna, Austria The mammalian kidney develops in three successive steps from the initial pronephros via the mesonephros to the adult metanephros. Although the nephric lineage is specified during pronephros induction, no single regulator, including the transcription factor Pax2 or Pax8, has yet been identified to control this initial phase of kidney development. In this paper, we demonstrate that mouse embryos lacking both Pax2 and Pax8 are unable to form the pronephros or any later nephric structures. In these double-mutant embryos, the intermediate mesoderm does not undergo the mesenchymal-epithelial transitions required for nephric duct formation, fails to initiate the kidney-specific expression of Lim1and c -Ret, and is lost by apoptosis 1d after failed pronephric induction. Conversely, retroviral misexpression of Pax2 was sufficient to induce ectopic nephric structures in the intermediate mesoderm and genital ridge of chick embryos. Together, these data identify Pax2 and Pax8 as critical regulators that specify the nephric lineage. [Keywords: Pax2;Pax8;genetic redundancy;kidney development;mesenchymal-epithelial transition;lineage specification] Received June 28, 2002;revised version accepted September 20, 2002. Kidney development in mammals and birds proceeds in development (for review, see Kuure et al. 2000;Davies three successive steps that are all characterized by the and Brändli 2002). The majority of these genes are essen- mesenchymal-to-epithelial transformation of intermedi- tial for proper morphogenesis of the metanephros with ate mesoderm cells. -
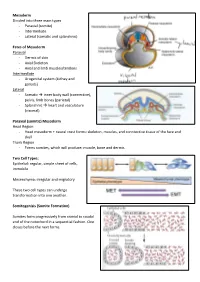
Mesoderm Divided Into Three Main Types - Paraxial (Somite) - Intermediate - Lateral (Somatic and Splanchnic)
Mesoderm Divided into three main types - Paraxial (somite) - Intermediate - Lateral (somatic and splanchnic) Fates of Mesoderm Paraxial - Dermis of skin - Axial Skeleton - Axial and limb muscles/tendons Intermediate - Urogenital system (kidney and gonads) Lateral - Somatic inner body wall (connective), pelvis, limb bones (parietal) - Splanchnic heart and vasculature (visceral) Paraxial (somitic) Mesoderm Head Region - Head mesoderm + neural crest forms: skeleton, muscles, and conntective tissue of the face and skull Trunk Region - Forms somites, which will produce: muscle, bone and dermis Two Cell Types: Epithelial: regular, simple sheet of cells, immobile Mesenchyma: irregular and migratory These two cell types can undergo transformation into one another. Somitogenisis (Somite Formation) Somites form progressively from cranial to caudal end of the notochord in a sequential fashion. One closes before the next forms. Somite Differentiation The somite splits into the epithelial dermamyotome (dermis/muscle) and the messenchymal sclerotome (skeletal). The somite is all paraxial mesoderm. Somite location determines the fate of its associates derma/myo/sclerotomes. Intermediate Mesoderm Urogenital system: - Kidneys - Gonads - Reproductive Duct Systems Runs alongside the paraxial mesoderm. Urogenital System Along mesonephric duct: - Pronephros, mesonephros, and metanephros - Pronephros fall away as gonad develops on ventral-medial side of mesonephros. - Metanephrogenic mesenchyme gives rise to kidney. The mesonephric duct will become the Wolffian duct forming at the nephric bud. The Mullerian duct forms via an invagination on the dorsal side of the nephric duct. The gonad will degenerate one of the two ducts depending on the hormones it produces. XX degenerates Wolffian duct – no testosterone, anti-Mullerian hormone (AMH) not produced, and Mullerian duct can develop in addition to female reproductive organs (ovaries, vagina) XY degenerates Mullerian duct – testosterone, AMH produced, Wolffian duct continues as male reproductive organs (testes, penis) develop. -

Renal Development
RENAL DEVELOPMENT Jon Barasch M.D., Ph.D. Telephone: 305-1890 e-mail: [email protected] SUGGESTED READING: Larsen, 3rd edition, pp 265 (first three paragraphs) - 266, 268-276 and figure 10-10 LEARNING OBJECTIVES: You should be able to: 1. Describe the three kidneys that are produced during development and know what happens to each one. 2. Explain what is meant by ‘reciprocal induction’ and why it poses problems in interpreting experiments in developing kidney. 3. Describe the stages of nephron formation from the renal vesicle. 4. Discuss the regulators of mesenchymal to epithelial transition in the intermediate mesoderm and metanephric mesenchyme and name three molecules mediating conversion. 5. Describe branching morphogenesis and name the three patterns in the developing metanephros. 6. Discuss three key important ligands and their receptors. 7. Discuss the classification of congenital renal abnormalities that are associated with urological abnormalities and the possible underlying mechanisms for their association. SUMMARY: The urogenital system derives from mesenchymal cells by a process of conversion to epithelia. The development of the kidney relies on three mechanisms of epithelial morphogenesis. 1. Some newborn epithelia migrate extensively (Wolffian duct), 2. some undergo branching morphogenesis (ureteric bud) and 3. some produce highly segmented tubules (nephrons). GLOSSARY: Angiotensin II: your favorite vasoconstrictor and regulator of proximal tubule reclamation of NaCl and water by receptor type 1. Receptor type-2 modulates cell growth and seems to play a role in congenital abnormalities. Arcade: a tubule of ureteric bud that induces a few nephrons simultaneously. The nephrons join to a common drainage called a connecting tubule that feeds into the ureteric’s collecting duct. -
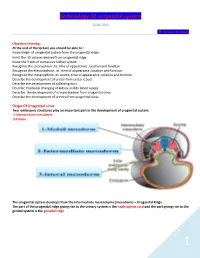
Embryology of Urogenital System
Embryology Of urogenital system 2018-2019 DR. Hassna B. Jawad Objective learning : At the end of the lecture you should be able to : Know Origin of urogenital system from the urogenital ridge Enlist the Structures derived from urogenital ridge Know the 3 sets of successive kidney system Recognize the pronephron ,its time of appearance ,location and function Recognize the Mesonephron, its time of appearance ,location and function Recognize the metanephron ,its source, time of appearance ,location and function Describe the development of ureter from ureteric bud Describe the development of collecting duct Describe Positional changing of kidney and its blood supply Describe the development of urinary bladder from urogenital sinus Describe the development of urethra from urogenital sinus Origin Of Urogenital sinus Two embryonic structures play an important part in the development of urogenital system: 1-Intermediate mesoderm 2-Cloaca The urogenital system develops from the intermediate mesenchyme (mesoderm) – Urogenital Ridge. The part of the urogenital ridge giving rise to the urinary system is the nephrogenic cord and the part giving rise to the genital system is the gonadal ridge. 1 Embryology Of urogenital system 2018-2019 DR. Hassna B. Jawad Development of urinary system: 3 sets of successive kidneys : Pronephron: These bilateral structures appear early in the fourth week. Segmented division of intermediate mesoderm form a few cell clusters and 5-7 pairs pronephric tubules in the cervical region. One end of the tubules opened at coelomic cavity and the other end opened in to pronephric duct 2 Embryology Of urogenital system 2018-2019 DR. Hassna B. Jawad Function : The tubules transmits the waste product from coelomic cavity to the pronephric duct that runs caudally and open into the Cloaca . -

Kidney Development and Function in the Fetus
Kidney Development, - and Function in the Fetus Bob Caruthers, CST, PhD The kidneys produce urine, a blood filtrate, and regulate uri- can be seen in the fourth week. The pronephros is complete1 nary volume and composition. These regulatory activities regred by the start of the fifth week. The pronephros form involve balancing water and solute transport, conserving and regresses in a cranial-to-caudal sequence. No pronephric nutrients, eliminating waste products, and regulating acid and glomeruli (cluster of capillaries) have been observed, and no bases. The primary purpose of kidney function is to maintain a vesicles are associated with the pronephric duct. The stable environment in which cellular and tissue metabolic pronephros is, therefore, not active in urine producti0n.Y activity can proceed at an optimal level. The kidneys secrete the hormone renin, erythropoietin and 1.25-dihydroxy vita- MPSONEPHROS min D. Renin helps regulate blood pressure. Erythropoietin The mesonephros originates in the nephrogenic cord that is helps regulate erythrocyte production. 1,25-dihydroxy vitamin part of the intermediate mesoderm. Early mesonephric forma- D plays a role in calcium metabolism. This article will discuss tion is evident before the pronephros has completed its regre: the development of the kidney, and its role in the fetus."' sion. The mesonephros also degenerates in a cranial-to-cauda Since the kidneys are bilateral structures, development sequence. Some of the cranial structures are degenerating in involves both right and left kidneys. During fetal develop- the fifth week of fetal development while the caudal structun ment, three separate nephric structures develop in succession; are still differentiating. -

Embryology and Teratology in the Curricula of Healthcare Courses
ANATOMICAL EDUCATION Eur. J. Anat. 21 (1): 77-91 (2017) Embryology and Teratology in the Curricula of Healthcare Courses Bernard J. Moxham 1, Hana Brichova 2, Elpida Emmanouil-Nikoloussi 3, Andy R.M. Chirculescu 4 1Cardiff School of Biosciences, Cardiff University, Museum Avenue, Cardiff CF10 3AX, Wales, United Kingdom and Department of Anatomy, St. George’s University, St George, Grenada, 2First Faculty of Medicine, Institute of Histology and Embryology, Charles University Prague, Albertov 4, 128 01 Prague 2, Czech Republic and Second Medical Facul- ty, Institute of Histology and Embryology, Charles University Prague, V Úvalu 84, 150 00 Prague 5 , Czech Republic, 3The School of Medicine, European University Cyprus, 6 Diogenous str, 2404 Engomi, P.O.Box 22006, 1516 Nicosia, Cyprus , 4Department of Morphological Sciences, Division of Anatomy, Faculty of Medicine, C. Davila University, Bucharest, Romania SUMMARY Key words: Anatomy – Embryology – Education – Syllabus – Medical – Dental – Healthcare Significant changes are occurring worldwide in courses for healthcare studies, including medicine INTRODUCTION and dentistry. Critical evaluation of the place, tim- ing, and content of components that can be collec- Embryology is a sub-discipline of developmental tively grouped as the anatomical sciences has biology that relates to life before birth. Teratology however yet to be adequately undertaken. Surveys (τέρατος (teratos) meaning ‘monster’ or ‘marvel’) of teaching hours for embryology in US and UK relates to abnormal development and congenital medical courses clearly demonstrate that a dra- abnormalities (i.e. morphofunctional impairments). matic decline in the importance of the subject is in Embryological studies are concerned essentially progress, in terms of both a decrease in the num- with the laws and mechanisms associated with ber of hours allocated within the medical course normal development (ontogenesis) from the stage and in relation to changes in pedagogic methodol- of the ovum until parturition and the end of intra- ogies.