Genetic Anomalies in Fetuses with Tetralogy of Fallot by Using High
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
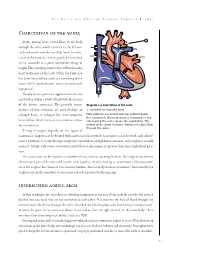
Coarctation of the Aorta Interrupted Aortic Arch
T HE P EDIATRIC C ARDIAC S URGERY I NQUEST R EPORT Coarctation of the aorta In the normal heart, blood flows to the body through the aorta, which connects to the left ven- tricle and arches over the top of the heart.In coarc- tation of the aorta,the aorta is pinched (in medical terms, coarcted) at a point somewhere along its length. This pinching restricts blood flow from the heart to the rest of the body. Often, the baby also has other heart defects, such as a narrowing of the aortic arch (in medical terms,transverse aortic arch hypoplasia). Usually no symptoms are apparent at birth, but can develop within a week of birth with the closure of the ductus arteriosus. The possible conse- Diagram 2.9 Coarctation of the aorta quences of this condition are poor feeding, an 1 – pinched or coarcted aorta enlarged heart, an enlarged liver and congestive Flow patterns are normal but are reduced below the coarctation. Blood pressure is increased in ves- heart failure. Blood pressure also increases above sels leaving the aorta above the coarctation. The the constriction. broken white arrow indicates diminished blood flow through the aorta. Timing of surgery depends on the degree of coarctation. Surgery may be delayed with mild coarctation (which sometimes is not detected until adoles- cence). However, if a baby develops congestive heart failure or high blood pressure, early surgery is usually required. A baby with severe coarctation should have early surgery to prevent long-term high blood pres- sure. The coarctation can be repaired in a number of ways without opening the heart.The surgeon can remove the narrowed part of the aorta and sew the ends together, thereby creating an anastomosis. -

Pulmonary-Atresia-Mapcas-Pavsdmapcas.Pdf
Normal Heart © 2012 The Children’s Heart Clinic NOTES: Children’s Heart Clinic, P.A., 2530 Chicago Avenue S, Ste 500, Minneapolis, MN 55404 West Metro: 612-813-8800 * East Metro: 651-220-8800 * Toll Free: 1-800-938-0301 * Fax: 612-813-8825 Children’s Minnesota, 2525 Chicago Avenue S, Minneapolis, MN 55404 West Metro: 612-813-6000 * East Metro: 651-220-6000 © 2012 The Children’s Heart Clinic Reviewed March 2019 Pulmonary Atresia, Ventricular Septal Defect and Major Aortopulmonary Collateral Arteries (PA/VSD/MAPCAs) Pulmonary atresia (PA), ventricular septal defect (VSD) and major aortopulmonary collateral arteries (MAPCAs) is a rare type of congenital heart defect, also referred to as Tetralogy of Fallot with PA/MAPCAs. Tetralogy of Fallot (TOF) is the most common cyanotic heart defect and occurs in 5-10% of all children with congenital heart disease. The classic description of TOF includes four cardiac abnormalities: overriding aorta, right ventricular hypertrophy (RVH), large perimembranous ventricular septal defect (VSD), and right ventricular outflow tract obstruction (RVOTO). About 20% of patients with TOF have PA at the infundibular or valvar level, which results in severe right ventricular outflow tract obstruction. PA means that the pulmonary valve is closed and not developed. When PA occurs, blood can not flow through the pulmonary arteries to the lungs. Instead, the child is dependent on a patent ductus arteriosus (PDA) or multiple systemic collateral vessels (MAPCAs) to deliver blood to the lungs for oxygenation. These MAPCAs usually arise from the de- scending aorta and subclavian arteries. Commonly, the pulmonary arteries are abnormal, with hypoplastic (small and underdeveloped) central and branch pulmonary arteries and/ or non-confluent central pulmonary arteries. -

Prenatal Diagnosis, Associated Findings and Postnatal Outcome Of
J. Perinat. Med. 2019; 47(3): 354–364 Ingo Gottschalka,*, Judith S. Abela, Tina Menzel, Ulrike Herberg, Johannes Breuer, Ulrich Gembruch, Annegret Geipel, Konrad Brockmeier, Christoph Berg and Brigitte Strizek Prenatal diagnosis, associated findings and postnatal outcome of fetuses with double outlet right ventricle (DORV) in a single center https://doi.org/10.1515/jpm-2018-0316 anomalies, 30 (66.7%) had extracardiac anomalies and 13 Received September 18, 2018; accepted November 26, 2018; (28.9%) had chromosomal or syndromal anomalies. Due previously published online December 20, 2018 to their complex additional anomalies, five (11.1%) of our Abstract 45 fetuses had multiple malformations and were highly suspicious for non-chromosomal genetic syndromes, Objective: To assess the spectrum of associated anoma- although molecular diagnosis could not be provided. Dis- lies, the intrauterine course, postnatal outcome and orders of laterality occurred in 10 (22.2%) fetuses. There management of fetuses with double outlet right ventricle were 17 terminations of pregnancy (37.8%), two (4.4%) (DORV). intrauterine and seven (15.6%) postnatal deaths. Nineteen Methods: All cases of DORV diagnosed prenatally over a of 22 (86.4%) live-born children with an intention to treat period of 8 years were retrospectively collected in a single were alive at last follow-up. The mean follow-up among tertiary referral center. All additional prenatal findings survivors was 32 months (range, 2–72). Of 21 children who were assessed and correlated with the outcome. The accu- had already undergone postnatal surgery, eight (38.1%) racy of prenatal diagnosis was assessed. achieved biventricular repair and 13 (61.9%) received Results: Forty-six cases of DORV were diagnosed pre- univentricular palliation. -

Double Outlet Right Ventricle (DORV)
Double outlet right ventricle (DORV) Vita Zīdere, MD FRCP Consultant Paediatric and Fetal Cardiologist • Complex lesion, represents 3 % of CHD seen in fetus • Both Great Arteries arise completely or predominantly from the morphological right ventricle • Associates with increased NT, genetic and extracardiac anomalies • Double outlet right ventricle is a spectrum of abnormalities • almost always has a VSD • heterogeneous with respect to size and position of VSD and relationship of GA • individual anatomy dictates surgical approach and outcome • It poses a number of challenges • Definition • Anatomic Variability (incl. normal or abnormal AV connections) • Surgical options DORV TOF type DORV “Simple” DORV Complex DORV Always VSD: Unbalanced Ventricles AVSD (often RAI or LAI) subaortic, MV atresia subpulmonary or uncommitted; Coarctation, IAA +/-PS Discordant AV connections Criss-cross heart Normal atrial situs Patent, concordant AV connections DORV Overriding aorta Both GA (parallel) from RV “50 % rule” Subpulmonary, subaortic or uncommitted VSD TOF type DORV Anatomical repair- Repair depends from GA relationship with VSD VSD closure & PS release LV to Ao baffle ASO with baffling VSD to neo-Ao Rarely BV repair is unfeasible L G Price 2004 G Price • True override is independent of septal axis • postnatally is assessed relative to chord of circle in short axis* *BR Wilcox, AC Cook, RH Anderson. Surgical anatomy of the heart,2004 Double Outlet Right ventricle v. Tetralogy of Fallot L L R R • Perimembranous VSD, overriding aorta • Pulmonary artery -

CYANOTIC CONGENITAL HEART DISEASE by JAMES W
.-51.I Postgrad Med J: first published as 10.1136/pgmj.25.289.511 on 1 November 1949. Downloaded from CYANOTIC CONGENITAL HEART DISEASE By JAMES W. BROWN, M.D., F.R.C.P. Physician, General Hospital, Grimsby, and Grimsby and Lindsey Rheumatism and Heart Clinics The last 15 years have witnessed a very re- it was possible to build up a clinical picture and so markable change in the attitude of the clinician arrive at a reasonably correct anatomical diagnosis. towards cyanotic congenital heart disease. It is At the same time knowledge of the natural history within the memory of many that it was the custom of congenital heart disease has also increased, and to make a simple diagnosis of congenital heart some idea of its prognosis has been ascertained. abnormality with cyanosis, and express the opinion Lastly, the conception of Taussig that an in- that little or nothing could be done about it. Then adequate pulmonary blood supply was the critical there appeared the pioneer work of Abbott and abnormality in many cyanotic cases, and the de- others which produced not only a classification of velopment of an anastomotic operation between the the various abnormalities, but furnished a mass of systemic and pulmonary circulations so as to clinical and postmortem observations from which furnish an artificial ductus arteriosus, by Blalock Protected by copyright. http://pmj.bmj.com/ on September 23, 2021 by guest. FIG. i.-Tetralogy of Fallot. Female aged 3 years. Right ventricle opened. An arrow marks the subvalvular stenosis. of the pulmonary artery. Ventricular septal defect with probe passing into it behind the crista supraventricularis. -

|||GET||| the Complete Reference for Scimitar Syndrome 1St Edition
THE COMPLETE REFERENCE FOR SCIMITAR SYNDROME 1ST EDITION DOWNLOAD FREE Vladimiro Vida | 9780128104071 | | | | | Scimitar syndrome: report of a case and its surgical management URL of The Complete Reference for Scimitar Syndrome 1st edition. Check for errors and try again. This is an advantage of the book. His major research areas include congenital heart disease in adults, congenital heart surgery, minimally invasive surgery, and Scimitar Syndrome. Pulmonary Development 3. Brand new color images to illustrate Functional imaging techniques. Covers both common and uncommon disorders. When you read an eBook on VitalSource Bookshelf, enjoy such features as: Access online or offline, on mobile or desktop devices Bookmarks, highlights and notes sync across all your devices Smart study tools such as note sharing and subscription, review mode, and Microsoft OneNote integration Search and navigate content across your entire Bookshelf library Interactive notebook and read-aloud functionality Look up additional information online by highlighting a word or phrase. By System:. We would like to ask you for a moment of your time to fill in a short questionnaire, at the end of your visit. J Card Surg. Access the full text online and download images via Expert Consult Access the latest version of the Fleischner Society's glossary of terms for thoracic imaging. We believe that in this case, the scimitar vein entered the hepatic venous system. Curving downward through the right lower lung field was a band-like shadow which widened gradually as it descended, paralleling the right cardiac border. At 4 years, there were no post-operative complications. Over time, this can lead to right heart dilation The Complete Reference for Scimitar Syndrome 1st edition pulmonary hypertension. -

Scimitar Syndrome with Tetralogy of Fallot and Pulmonary Atresia
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector The Egyptian Heart Journal (2015) 67, 275–277 HOSTED BY Egyptian Society of Cardiology The Egyptian Heart Journal www.elsevier.com/locate/ehj www.sciencedirect.com CASE REPORT Scimitar syndrome with tetralogy of fallot and pulmonary atresia Sameh R. Ismail *, Mustafa A. Al-Muhaya, Mohamed S. Kabbani King Abdulaziz Medical City, King Saud University for Health Sciences, Department of Cardiac Sciences, National Guard hospital Health Affairs, Riyadh, Saudi Arabia Received 2 June 2014; accepted 27 August 2014 Available online 18 September 2014 KEYWORDS Abstract Scimitar syndrome is a rare variant of partial anomalous pulmonary venous connection. Scimitar syndrome; The association of Scimitar syndrome with another cardiac congenital anomaly such as tetralogy of Tetralogy of fallot; Fallot with pulmonary atresia is extremely rare; we are reporting a successful combined treatment Pulmonary atresia using transcatheter closure of major aorto-pulmonary collateral and a single-stage surgical correction in eighteen month old boy diagnosed as Scimitar syndrome with tetralogy of Fallot and pulmonary atresia. ª 2014 Production and hosting by Elsevier B.V. on behalf of Egyptian Society of Cardiology. 1. Case report arteriosus, bilateral superior vena cava and partial anomalous pulmonary venous drainage (PAPVD). The right pulmonary An eighteen month old boy was referred to our center due to veins are draining to the inferior vena cava–right atrium cyanosis and shortness of breath. He was a product of full term (IVC–RA) junction (Fig. 1). In order to confirm the diagnosis normal vaginal delivery and the first child in his family. -

Tetralogy of Fallot
Department Of Health and Mental Hygiene Prevention and Health Promotion Administration Office for Genetics and People with Special Health Care Needs Tetralogy of Fallot What is Tetralogy of Fallot? Heart defect which is present at birth (congenital) Most common type of critical congenital heart defect Consists of four defects of the heart and the major blood vessels: 1. Ventricular Septal Defect (VSD) - a hole between the right and left ventricles (bottom chambers) of the heart 2. Narrowing of the pulmonary outflow tract - the valve and artery that connects the heart with the lungs 3. Overriding Aorta - aorta usually takes oxygen rich blood to the body from the left ventricle to the body. An overriding aorta is shifted and takes blood from the right and left ventricles, reducing the amount of oxygen in the blood that goes to the body 4. Right Ventricular Hypertrophy - thickened wall of the right ventricle What is the cause of Tetralogy of Fallot? There is no known cause, but there are certain risk factors which will increase the chance of this condition occurring: 1. Drinking alcohol regularly during pregnancy 2. Maternal diabetes 3. Mother’s age (over 40) 4. Poor nutrition during pregnancy 5. Rubella or other viral illnesses during pregnancy Infants born with Tetralogy of Fallot are more likely to have chromosomal (genetic) disorders such as Down Syndrome or DiGeorge Syndrome Signs and Symptoms Bluish skin color, especially around lips and fingernails Clubbing of fingers (broadening and blunting of tips) Poor feeding and poor growth Fainting episodes How is Tetralogy of Fallot treated? Medicines are often used to improve blood flow and circulation. -

Tetralogy of Fallot: Basic Imaging Findings and Management
Journal of Radiology Nursing 38 (2019) 164e167 Contents lists available at ScienceDirect Journal of Radiology Nursing journal homepage: www.sciencedirect.com/journal/ journal-of-radiology-nursing Tetralogy of Fallot: Basic Imaging Findings and Management * Ramya Vajapey, MD, David Majdalany, MD Division of Clinical Cardiology, Department of Heart and Vascular Institute, Cleveland Clinic Foundation, Cleveland, Ohio abstract Keywords: Tetralogy of Fallot (TOF) is one of the most common cyanotic, congenital heart disease with complex Tetralogy of Fallot anatomic malformations and unknown etiology. With medical advancements, most patients in this Congenital cohort require early total correction and survive into adulthood. This review addresses various imaging Malformation modalities that help diagnose TOF, methods to correct and repair TOF-associated cardiac malformations, Right ventricular hypertrophy and the sequelae and long-term complications of this disease. © 2019 Association for Radiologic & Imaging Nursing. Published by Elsevier Inc. All rights reserved. Background Imaging findings Tetralogy of Fallot (TOF) is one of the most common causes of Electrocardiogram/Chest X-ray cyanotic cardiac disease and occurs in 3 of every 10,000 births, ac- counting for about 10% of all congenital heart diseases. TOF is a Initial evaluation of TOF includes obtaining an electrocardio- cardiac malformation that has a ventricular septal defect, obstruc- gram, which will typically demonstrate right axis deviation with tion of right ventricular outflow tract, aortic root overriding the right bundle branch block and right ventricular hypertrophy with ventricular septum, and right ventricular hypertrophy (see Figure 1) large R waves in anterior precordial leads and large S wave in the (Bailliard & Anderson, 2009). It was first described by Danish lateral precordial leads. -

Early Embryonic Expression of AP-2 Is Critical for Cardiovascular Development
Journal of Cardiovascular Development and Disease Article Early Embryonic Expression of AP-2α Is Critical for Cardiovascular Development Amy-Leigh Johnson 1, Jürgen E. Schneider 2, Timothy J. Mohun 3, Trevor Williams 4, Shoumo Bhattacharya 5 , Deborah J. Henderson 1, Helen M. Phillips 1 and Simon D. Bamforth 1,* 1 Newcastle University Biosciences Institute, Centre for Life, Newcastle NE1 3BZ, UK; [email protected] (A.-L.J.); [email protected] (D.J.H.); [email protected] (H.M.P.) 2 Biomedical Imaging, University of Leeds, Leeds LS2 9JT, UK; [email protected] 3 The Francis Crick Institute, London NW1 1AT, UK; [email protected] 4 Department of Craniofacial Biology, University of Colorado Anshutz Medical Campus, Aurora, CO 80045, USA; [email protected] 5 Department of Cardiovascular Medicine, University of Oxford, Wellcome Trust Centre for Human Genetics, Oxford OX3 7BN, UK; [email protected] * Correspondence: [email protected]; Tel.: +44-191-241-8764 Received: 22 June 2020; Accepted: 22 July 2020; Published: 23 July 2020 Abstract: Congenital cardiovascular malformation is a common birth defect incorporating abnormalities of the outflow tract and aortic arch arteries, and mice deficient in the transcription factor AP-2α (Tcfap2a) present with complex defects affecting these structures. AP-2α is expressed in the pharyngeal surface ectoderm and neural crest at mid-embryogenesis in the mouse, but the precise tissue compartment in which AP-2α is required for cardiovascular development has not been identified. In this study we describe the fully penetrant AP-2α deficient cardiovascular phenotype on a C57Bl/6J genetic background and show that this is associated with increased apoptosis in the pharyngeal ectoderm. -

Scimitar Syndrome with Tetralogy of Fallot and Pulmonary Atresia
The Egyptian Heart Journal (2014) xxx, xxx–xxx HOSTED BY Egyptian Society of Cardiology The Egyptian Heart Journal www.elsevier.com/locate/ehj www.sciencedirect.com SHORT COMMUNICATION Scimitar syndrome with tetralogy of fallot and pulmonary atresia Sameh R. Ismail *, Mustafa A. Al-Muhaya, Mohamed S. Kabbani King Abdulaziz Medical City, King Saud University for Health Sciences, Department of Cardiac Sciences, National Guard hospital Health Affairs, Riyadh, Saudi Arabia Received 2 June 2014; accepted 27 August 2014 KEYWORDS Abstract Scimitar syndrome is a rare variant of partial anomalous pulmonary venous connection. Scimitar syndrome; The association of Scimitar syndrome with another cardiac congenital anomaly such as tetralogy of Tetralogy of fallot; Fallot with pulmonary atresia is extremely rare; we are reporting a successful combined treatment Pulmonary atresia using transcatheter closure of major aorto-pulmonary collateral and a single-stage surgical correction in eighteen month old boy diagnosed as Scimitar syndrome with tetralogy of Fallot and pulmonary atresia. ª 2014 Production and hosting by Elsevier B.V. on behalf of Egyptian Society of Cardiology. 1. Case report arteriosus, bilateral superior vena cava and partial anomalous pulmonary venous drainage (PAPVD). The right pulmonary An eighteen month old boy was referred to our center due to veins are draining to the inferior vena cava–right atrium cyanosis and shortness of breath. He was a product of full term (IVC–RA) junction (Fig. 1). In order to confirm the diagnosis normal vaginal delivery and the first child in his family. His Cardiac Computed Tomography and Angiography (CT angio- birth weight was 3.5 kg. On admission his weight was 14 kg gram) was performed, it confirmed that the right pulmonary (95% centile) and height 83 cm (95% centile), oxygen satura- veins are draining to the IVC–RA junction above the dia- tion on room air 75–80%. -

Isolated Fetal Cardiac Abnormalities: Are They Really Isolated?
THIEME Case Report e355 Isolated Fetal Cardiac Abnormalities: Are They Really Isolated? Armin S. Razavi, MD1 Stephen T. Chasen, MD1 1 Maternal-Fetal Medicine, Department of Obstetrics and Gynecology, Address for correspondence Armin S. Razavi, MD, Maternal-Fetal Weill Cornell Medicine, New York, New York Medicine, Department of Obstetrics and Gynecology, Weill Cornell Medicine, 525 East 68th Street, Box 122, New York, NY 10065 Am J Perinatol Rep 2018;8:e355–e358. (e-mail: [email protected]). Abstract Objective To determine the rate of unsuspected noncardiac abnormalities in new- borns suspected to have isolated cardiac abnormalities in the second trimester. Study Design A review of the ultrasound database from the Weill Cornell Medical Center identified fetuses with a suspected cardiac abnormality from January 2006 to November 2016. Cases with prenatally suspected noncardiac structural abnormalities, abnormal fetal or neonatal karyotype or microarray, and those who delivered at an outside institution or underwent abortion were excluded. Neonatal records were reviewed to confirm prenatal findings and to identify anomalies not suspected in the second trimester. Results Sixty-eight live births met the inclusion criteria. Five newborns (7.4%) had major abnormalities not identified in the second trimester. Three newborns had an imperforate anus. One newborn had left hydronephrosis and absent right lung, and one Keywords had hemifacial microsomia and fused ribs. All five newborns with unsuspected ► cardiac abnormality anomalies were in the group with suspected conotruncal anomalies, with a 11.9% ► cardiac anomaly rate of unsuspected anomalies versus 0% in those with nonconotruncal cardiac ► conotruncal anomalies (p ¼ 0.15). ► noncardiac Conclusion Patients with a suspected isolated fetal cardiac anomaly on ultrasound abnormalities should be aware of the possibility of other major structural abnormalities, especially in ► noncardiac anomalies cases of conotruncal cardiac anomalies.