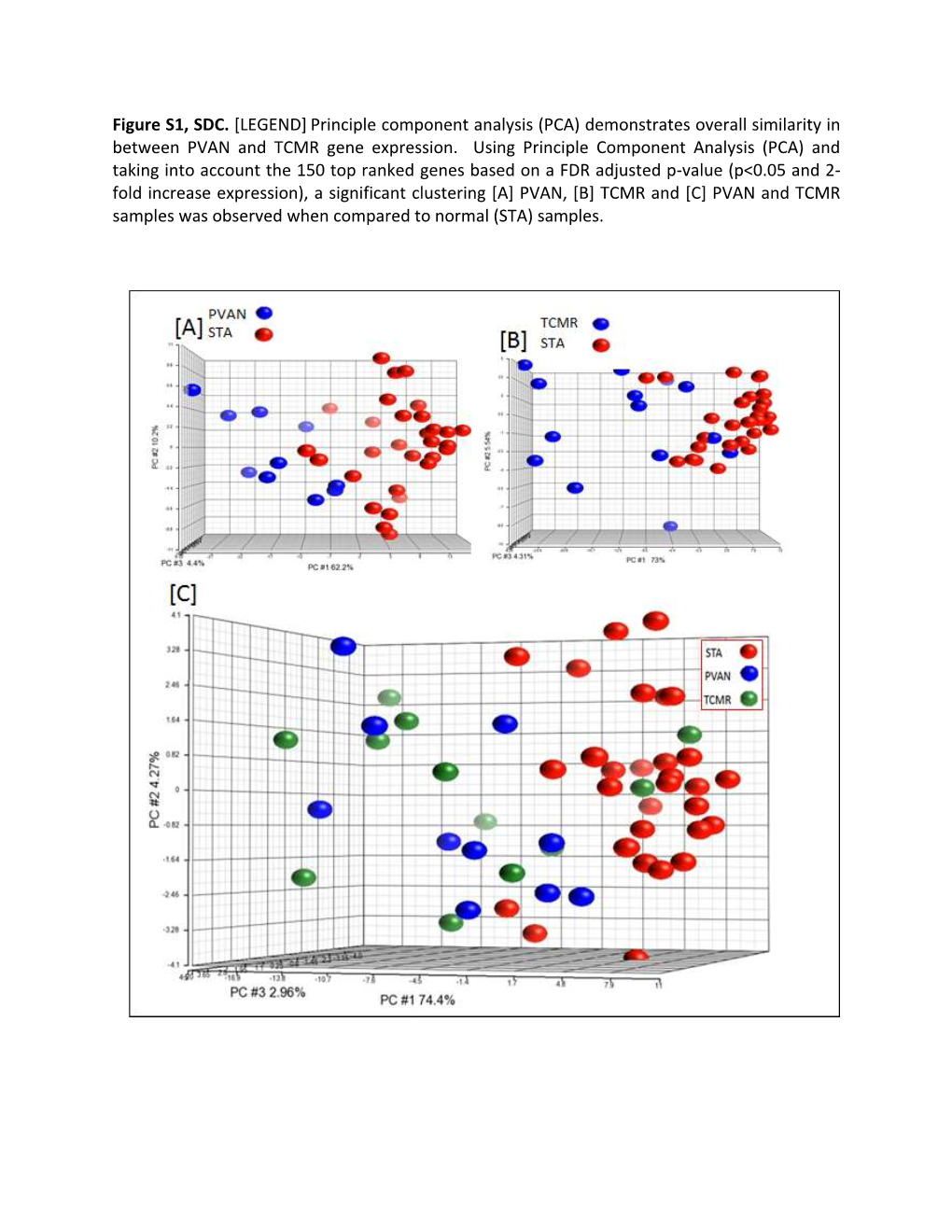
Figure S1, SDC. [LEGEND] Principle Component Analysis (PCA) Demonstrates Overall Similarity in Between PVAN and TCMR Gene Expression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Viewed Under 23 (B) Or 203 (C) fi M M Male Cko Mice, and Largely Unaffected Magni Cation; Scale Bars, 500 M (B) and 50 M (C)
BRIEF COMMUNICATION www.jasn.org Renal Fanconi Syndrome and Hypophosphatemic Rickets in the Absence of Xenotropic and Polytropic Retroviral Receptor in the Nephron Camille Ansermet,* Matthias B. Moor,* Gabriel Centeno,* Muriel Auberson,* † † ‡ Dorothy Zhang Hu, Roland Baron, Svetlana Nikolaeva,* Barbara Haenzi,* | Natalya Katanaeva,* Ivan Gautschi,* Vladimir Katanaev,*§ Samuel Rotman, Robert Koesters,¶ †† Laurent Schild,* Sylvain Pradervand,** Olivier Bonny,* and Dmitri Firsov* BRIEF COMMUNICATION *Department of Pharmacology and Toxicology and **Genomic Technologies Facility, University of Lausanne, Lausanne, Switzerland; †Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts; ‡Institute of Evolutionary Physiology and Biochemistry, St. Petersburg, Russia; §School of Biomedicine, Far Eastern Federal University, Vladivostok, Russia; |Services of Pathology and ††Nephrology, Department of Medicine, University Hospital of Lausanne, Lausanne, Switzerland; and ¶Université Pierre et Marie Curie, Paris, France ABSTRACT Tight control of extracellular and intracellular inorganic phosphate (Pi) levels is crit- leaves.4 Most recently, Legati et al. have ical to most biochemical and physiologic processes. Urinary Pi is freely filtered at the shown an association between genetic kidney glomerulus and is reabsorbed in the renal tubule by the action of the apical polymorphisms in Xpr1 and primary fa- sodium-dependent phosphate transporters, NaPi-IIa/NaPi-IIc/Pit2. However, the milial brain calcification disorder.5 How- molecular identity of the protein(s) participating in the basolateral Pi efflux remains ever, the role of XPR1 in the maintenance unknown. Evidence has suggested that xenotropic and polytropic retroviral recep- of Pi homeostasis remains unknown. Here, tor 1 (XPR1) might be involved in this process. Here, we show that conditional in- we addressed this issue in mice deficient for activation of Xpr1 in the renal tubule in mice resulted in impaired renal Pi Xpr1 in the nephron. -

CCDC109B (MCUB) (NM 017918) Human Untagged Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for SC327798 CCDC109B (MCUB) (NM_017918) Human Untagged Clone Product data: Product Type: Expression Plasmids Product Name: CCDC109B (MCUB) (NM_017918) Human Untagged Clone Tag: Tag Free Symbol: MCUB Synonyms: CCDC109B Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin Fully Sequenced ORF: >OriGene SC327798 ORF sequence for NM_017918, the custom clone sequence may differ by one or more nucleotides ATGTTGTCAACAGTTGGTTCATTCCTTCAGGACCTACAAAATGAAGATAAGGGTATCAAAACTGCAGCCA TCTTCACAGCAGATGGCAACATGATTTCAGCTTCTACCTTGATGGATATTTTGCTAATGAATGATTTTAA ACTTGTCATTAATAAAATAGCATATGATGTGCAGTGTCCAAAGAGAGAAAAACCAAGTAATGAGCACACT GCTGAGATGGAACACATGAAATCCTTGGTTCACAGACTATTTACAATCTTGCATTTAGAAGAGTCTCAGA AAAAGAGAGAGCACCATTTACTGGAGAAAATTGACCACCTGAAGGAACAGCTGCAGCCCCTTGAACAGGT GAAAGCTGGAATAGAAGCTCATTCGGAAGCCAAAACCAGTGGACTCCTGTGGGCTGGATTGGCACTGCTG TCCATTCAGGGTGGGGCACTGGCCTGGCTCACGTGGTGGGTGTACTCCTGGGATATCATGGAGCCAGTTA CATACTTCATCACATTTGCAAATTCTATGGTCTTTTTTGCATACTTTATAGTCACTCGACAGGATTATAC TTACTCAGCTGTTAAGAGTAGGCAATTTCTTCAGTTCTTCCACAAGAAATCAAAGCAACAGCACTTTGAT GTGCAGCAATACAACAAGTTAAAAGAAGACCTTGCTAAGGCTAAAGAATCCCTGAAACAGGCGCGTCATT CTCTCTGTTTGCAAATGCAAGTAGAAGAACTCAATGAAAAGAATTAA Restriction Sites: SgfI-MluI ACCN: NM_017918 OTI Disclaimer: Our molecular clone sequence data has been matched to the reference identifier above as a point -

No Evidence of Association Between Complement Factor I Genetic Variant
European Journal of Human Genetics (2012) 20, 1–3 & 2012 Macmillan Publishers Limited All rights reserved 1018-4813/12 www.nature.com/ejhg LETTERS US-based sample of around 1200 cases with advanced AMD and No evidence of association 800 controls. The association signal extended over a region of about 175 kb, the most associated variant (Po10À7)beingtheSNP rs10033900 near the complement factor I (CFI) gene. Two replication between complement studies2,3 published also in this journal provided some additional support for an AMD susceptibility locus in this region. In the course factor I genetic variant of candidate gene studies of AMD, we had previously investigated SNPs spanning CFI including rs10033900 in a UK case–control rs10033900 and sample, which shows the expected associations with the well- established AMD-susceptibility loci CFH, ARMS2, CFB and C3.No age-related macular evidence of association with the CFI variants was observed. Following publication of the reports cited above we have typed rs10033900 degeneration in additional cases and controls in two independent samples from England and Scotland to investigate this further. Full details of the phenotyping criteria have been reported pre- viously.4 The English sample comprised of 859 cases with predomi- European Journal of Human Genetics (2012) 20, 1–2; nantly advanced AMD, either geographic atrophy (GA) or choroidal doi:10.1038/ejhg.2011.118; published online 12 October 2011 neovascularisation (CNV) and 423 examined controls. The Scottish sample consisted of 505 cases with either intermediate disease (age-related maculopathy, ARM) or advanced AMD, and 351 exam- In 2008, an association between age-related macular degeneration ined controls. -

Stelios Pavlidis3, Matthew Loza3, Fred Baribaud3, Anthony
Supplementary Data Th2 and non-Th2 molecular phenotypes of asthma using sputum transcriptomics in UBIOPRED Chih-Hsi Scott Kuo1.2, Stelios Pavlidis3, Matthew Loza3, Fred Baribaud3, Anthony Rowe3, Iaonnis Pandis2, Ana Sousa4, Julie Corfield5, Ratko Djukanovic6, Rene 7 7 8 2 1† Lutter , Peter J. Sterk , Charles Auffray , Yike Guo , Ian M. Adcock & Kian Fan 1†* # Chung on behalf of the U-BIOPRED consortium project team 1Airways Disease, National Heart & Lung Institute, Imperial College London, & Biomedical Research Unit, Biomedical Research Unit, Royal Brompton & Harefield NHS Trust, London, United Kingdom; 2Department of Computing & Data Science Institute, Imperial College London, United Kingdom; 3Janssen Research and Development, High Wycombe, Buckinghamshire, United Kingdom; 4Respiratory Therapeutic Unit, GSK, Stockley Park, United Kingdom; 5AstraZeneca R&D Molndal, Sweden and Areteva R&D, Nottingham, United Kingdom; 6Faculty of Medicine, Southampton University, Southampton, United Kingdom; 7Faculty of Medicine, University of Amsterdam, Amsterdam, Netherlands; 8European Institute for Systems Biology and Medicine, CNRS-ENS-UCBL, Université de Lyon, France. †Contributed equally #Consortium project team members are listed under Supplementary 1 Materials *To whom correspondence should be addressed: [email protected] 2 List of the U-BIOPRED Consortium project team members Uruj Hoda & Christos Rossios, Airways Disease, National Heart & Lung Institute, Imperial College London, UK & Biomedical Research Unit, Biomedical Research Unit, Royal -

Primate Specific Retrotransposons, Svas, in the Evolution of Networks That Alter Brain Function
Title: Primate specific retrotransposons, SVAs, in the evolution of networks that alter brain function. Olga Vasieva1*, Sultan Cetiner1, Abigail Savage2, Gerald G. Schumann3, Vivien J Bubb2, John P Quinn2*, 1 Institute of Integrative Biology, University of Liverpool, Liverpool, L69 7ZB, U.K 2 Department of Molecular and Clinical Pharmacology, Institute of Translational Medicine, The University of Liverpool, Liverpool L69 3BX, UK 3 Division of Medical Biotechnology, Paul-Ehrlich-Institut, Langen, D-63225 Germany *. Corresponding author Olga Vasieva: Institute of Integrative Biology, Department of Comparative genomics, University of Liverpool, Liverpool, L69 7ZB, [email protected] ; Tel: (+44) 151 795 4456; FAX:(+44) 151 795 4406 John Quinn: Department of Molecular and Clinical Pharmacology, Institute of Translational Medicine, The University of Liverpool, Liverpool L69 3BX, UK, [email protected]; Tel: (+44) 151 794 5498. Key words: SVA, trans-mobilisation, behaviour, brain, evolution, psychiatric disorders 1 Abstract The hominid-specific non-LTR retrotransposon termed SINE–VNTR–Alu (SVA) is the youngest of the transposable elements in the human genome. The propagation of the most ancient SVA type A took place about 13.5 Myrs ago, and the youngest SVA types appeared in the human genome after the chimpanzee divergence. Functional enrichment analysis of genes associated with SVA insertions demonstrated their strong link to multiple ontological categories attributed to brain function and the disorders. SVA types that expanded their presence in the human genome at different stages of hominoid life history were also associated with progressively evolving behavioural features that indicated a potential impact of SVA propagation on a cognitive ability of a modern human. -

1 Supporting Information for a Microrna Network Regulates
Supporting Information for A microRNA Network Regulates Expression and Biosynthesis of CFTR and CFTR-ΔF508 Shyam Ramachandrana,b, Philip H. Karpc, Peng Jiangc, Lynda S. Ostedgaardc, Amy E. Walza, John T. Fishere, Shaf Keshavjeeh, Kim A. Lennoxi, Ashley M. Jacobii, Scott D. Rosei, Mark A. Behlkei, Michael J. Welshb,c,d,g, Yi Xingb,c,f, Paul B. McCray Jr.a,b,c Author Affiliations: Department of Pediatricsa, Interdisciplinary Program in Geneticsb, Departments of Internal Medicinec, Molecular Physiology and Biophysicsd, Anatomy and Cell Biologye, Biomedical Engineeringf, Howard Hughes Medical Instituteg, Carver College of Medicine, University of Iowa, Iowa City, IA-52242 Division of Thoracic Surgeryh, Toronto General Hospital, University Health Network, University of Toronto, Toronto, Canada-M5G 2C4 Integrated DNA Technologiesi, Coralville, IA-52241 To whom correspondence should be addressed: Email: [email protected] (M.J.W.); yi- [email protected] (Y.X.); Email: [email protected] (P.B.M.) This PDF file includes: Materials and Methods References Fig. S1. miR-138 regulates SIN3A in a dose-dependent and site-specific manner. Fig. S2. miR-138 regulates endogenous SIN3A protein expression. Fig. S3. miR-138 regulates endogenous CFTR protein expression in Calu-3 cells. Fig. S4. miR-138 regulates endogenous CFTR protein expression in primary human airway epithelia. Fig. S5. miR-138 regulates CFTR expression in HeLa cells. Fig. S6. miR-138 regulates CFTR expression in HEK293T cells. Fig. S7. HeLa cells exhibit CFTR channel activity. Fig. S8. miR-138 improves CFTR processing. Fig. S9. miR-138 improves CFTR-ΔF508 processing. Fig. S10. SIN3A inhibition yields partial rescue of Cl- transport in CF epithelia. -

The Complex SNP and CNV Genetic Architecture of the Increased Risk of Congenital Heart Defects in Down Syndrome
Downloaded from genome.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press Research The complex SNP and CNV genetic architecture of the increased risk of congenital heart defects in Down syndrome M. Reza Sailani,1,2 Periklis Makrythanasis,1 Armand Valsesia,3,4,5 Federico A. Santoni,1 Samuel Deutsch,1 Konstantin Popadin,1 Christelle Borel,1 Eugenia Migliavacca,1 Andrew J. Sharp,1,20 Genevieve Duriaux Sail,1 Emilie Falconnet,1 Kelly Rabionet,6,7,8 Clara Serra-Juhe´,7,9 Stefano Vicari,10 Daniela Laux,11 Yann Grattau,12 Guy Dembour,13 Andre Megarbane,12,14 Renaud Touraine,15 Samantha Stora,12 Sofia Kitsiou,16 Helena Fryssira,16 Chariklia Chatzisevastou-Loukidou,16 Emmanouel Kanavakis,16 Giuseppe Merla,17 Damien Bonnet,11 Luis A. Pe´rez-Jurado,7,9 Xavier Estivill,6,7,8 Jean M. Delabar,18 and Stylianos E. Antonarakis1,2,19,21 1–19[Author affiliations appear at the end of the paper.] Congenital heart defect (CHD) occurs in 40% of Down syndrome (DS) cases. While carrying three copies of chromosome 21 increases the risk for CHD, trisomy 21 itself is not sufficient to cause CHD. Thus, additional genetic variation and/or environmental factors could contribute to the CHD risk. Here we report genomic variations that in concert with trisomy 21, determine the risk for CHD in DS. This case-control GWAS includes 187 DS with CHD (AVSD = 69, ASD = 53, VSD = 65) as cases, and 151 DS without CHD as controls. Chromosome 21–specific association studies revealed rs2832616 and rs1943950 as CHD risk alleles (adjusted genotypic P-values <0.05). -

“The Impact of ART on Genome‐Wide Oxidation of 5‐Methylcytosine and the Transcriptome During Early Mouse Development”
“The impact of ART on genome‐wide oxidation of 5‐methylcytosine and the transcriptome during early mouse development” Dissertation zur Erlangung des Grades “Doktor der Naturwissenschaften” am Fachbereich Biologie der Johannes Gutenberg-Universität Mainz Elif Diken geb. Söğütcü geb. am 22.07.1987 in Giresun-TURKEY Mainz 2016 Dekan: 1. Berichterstatter: 2. Berichterstatter: Tag der mündlichen Prüfung: Summary Summary The use of assisted reproductive technologies (ART) has been increasing over the past three decades due to the elevated frequency of infertility problems. Other factors such as easier access to medical aid than in the past and its coverage by health insurance companies in many developed countries also contributed to this growing interest. Nevertheless, a negative impact of ART on transcriptome and methylation reprogramming is heavily discussed. Methylation reprogramming directly after fertilization manifests itself as genome-wide DNA demethylation associated with the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in the pronuclei of mouse zygotes. To investigate the possible impact of ART particularly on this process and the transcriptome in general, pronuclear stage mouse embryos obtained upon spontaneous ovulation or superovulation through hormone stimulation representing ART were subjected to various epigenetic analyses. A whole- transcriptome RNA-Seq analysis of pronuclear stage embryos from spontaneous and superovulated matings demonstrated altered expression of the Bbs12 gene known to be linked to Bardet-Biedl syndrome (BBS) as well as the Dhx16 gene whose zebrafish ortholog was reported to be a maternal effect gene. Immunofluorescence staining with antibodies against 5mC and 5hmC showed that pronuclear stage embryos obtained by superovulation have an increased incidence of abnormal methylation and hydroxymethylation patterns in both maternal and paternal pronuclear DNA compared to their spontaneously ovulated counterparts. -

© Copyright 2021 Heather Raquel Dahlin
© Copyright 2021 Heather Raquel Dahlin The Structure of Sperm Autoantigenic Protein (SPA17): An R2D2 Protein Critical to Cilia and Implicated in Oncogenesis Heather Raquel Dahlin A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Washington 2021 Reading Committee: John D. Scott, Chair Ning Zheng Linda Wordeman Program Authorized to Offer Degree: Pharmacology University of Washington Abstract Structure of SPA17: An R2D2 Protein Critical to Cilia and Implicated in Oncogenesis Heather Raquel Dahlin Chair of the Supervisory Committee: John D. Scott, Ph.D., Edwin G. Krebs- Speights Professor of Cell Signaling and Cancer Biology Pharmacology A-Kinase Anchoring proteins (AKAPs) localize the activity of cyclic AMP (cAMP)-Dependent Protein Kinase (PKA) through interaction of an amphipathic helix that binds to a conserved RIIα docking and dimerization (R2D2) domain on the N-terminus of PKA. Genome analysis indicates that at least thirteen other RIIα superfamily proteins exist in humans, which are not coupled to cyclic nucleotide binding domains and are largely localized to cilia and flagella. The newly reported R2D2 proteins exist in two lineages differing by their similarity to Type I or Type II PKA. Moreover, R2D2 domains bind to AKAPs and can contain extra regulatory sequences conferring novel functions and binding specificity. Here we detail the structure of one such domain comprising the N-terminus of Sperm Autoantigenic Protein 17 (SPA17) resolved to 1.72 Å. The structure of core hydrophobic sites for dimerization and AKAP binding are highly conserved between PKA and SPA17. Additional flanking sequences outside of the core R2D2 domain occlude the AKAP binding site and reduce the affinity for AKAP helices in the absence of heterodimerization with another R2D2 protein, ROPN1L. -

Curcumin Alters Gene Expression-Associated DNA Damage, Cell Cycle, Cell Survival and Cell Migration and Invasion in NCI-H460 Human Lung Cancer Cells in Vitro
ONCOLOGY REPORTS 34: 1853-1874, 2015 Curcumin alters gene expression-associated DNA damage, cell cycle, cell survival and cell migration and invasion in NCI-H460 human lung cancer cells in vitro I-TSANG CHIANG1,2, WEI-SHU WANG3, HSIN-CHUNG LIU4, SU-TSO YANG5, NOU-YING TANG6 and JING-GUNG CHUNG4,7 1Department of Radiation Oncology, National Yang‑Ming University Hospital, Yilan 260; 2Department of Radiological Technology, Central Taiwan University of Science and Technology, Taichung 40601; 3Department of Internal Medicine, National Yang‑Ming University Hospital, Yilan 260; 4Department of Biological Science and Technology, China Medical University, Taichung 404; 5Department of Radiology, China Medical University Hospital, Taichung 404; 6Graduate Institute of Chinese Medicine, China Medical University, Taichung 404; 7Department of Biotechnology, Asia University, Taichung 404, Taiwan, R.O.C. Received March 31, 2015; Accepted June 26, 2015 DOI: 10.3892/or.2015.4159 Abstract. Lung cancer is the most common cause of cancer CARD6, ID1 and ID2 genes, associated with cell survival and mortality and new cases are on the increase worldwide. the BRMS1L, associated with cell migration and invasion. However, the treatment of lung cancer remains unsatisfactory. Additionally, 59 downregulated genes exhibited a >4-fold Curcumin has been shown to induce cell death in many human change, including the DDIT3 gene, associated with DNA cancer cells, including human lung cancer cells. However, the damage; while 97 genes had a >3- to 4-fold change including the effects of curcumin on genetic mechanisms associated with DDIT4 gene, associated with DNA damage; the CCPG1 gene, these actions remain unclear. Curcumin (2 µM) was added associated with cell cycle and 321 genes with a >2- to 3-fold to NCI-H460 human lung cancer cells and the cells were including the GADD45A and CGREF1 genes, associated with incubated for 24 h. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -

Evaluation of a New Method for Large-Scale and Gene-Targeted Next Generation DNA Sequencing in Nonmodel Species
University of Montana ScholarWorks at University of Montana Graduate Student Theses, Dissertations, & Professional Papers Graduate School 2013 Evaluation of a New Method for Large-Scale and Gene-targeted Next Generation DNA Sequencing in Nonmodel Species Ted Cosart The University of Montana Follow this and additional works at: https://scholarworks.umt.edu/etd Let us know how access to this document benefits ou.y Recommended Citation Cosart, Ted, "Evaluation of a New Method for Large-Scale and Gene-targeted Next Generation DNA Sequencing in Nonmodel Species" (2013). Graduate Student Theses, Dissertations, & Professional Papers. 4133. https://scholarworks.umt.edu/etd/4133 This Dissertation is brought to you for free and open access by the Graduate School at ScholarWorks at University of Montana. It has been accepted for inclusion in Graduate Student Theses, Dissertations, & Professional Papers by an authorized administrator of ScholarWorks at University of Montana. For more information, please contact [email protected]. EVALUTATION OF A NEW METHOD FOR LARGE-SCALE AND GENE- TARGETED NEXT GENERATION DNA SEQUENCING IN NONMODEL SPECIES By Ted Cosart BA, University of Montana, Missoula, Montana, 1983 MS, University of Montana, Missoula, Montana, 2006 Dissertation presented in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Individualized, Interdisciplinary Graduate Program The University of Montana Missoula, Montana August, 2013 Approved by: Sandy Ross, Associate Dean of The Graduate School Graduate School Dr. Jesse Johnson, Co-Chair Computer Science Dr. Gordon Luikart, Co-Chair Flathead Biological Station Dr. Jeffrey Good Division of Biological Sciences Dr. William Holben Division of Biological Sciences Dr. Stephen Porcella Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases Dr.