“The Impact of ART on Genome‐Wide Oxidation of 5‐Methylcytosine and the Transcriptome During Early Mouse Development”
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Molecular and Genetic Analysis of Otosclerosis
A molecular and genetic analysis of otosclerosis Joanna Lauren Ziff Submitted for the degree of PhD University College London January 2014 1 Declaration I, Joanna Ziff, confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. Where work has been conducted by other members of our laboratory, this has been indicated by an appropriate reference. 2 Abstract Otosclerosis is a common form of conductive hearing loss. It is characterised by abnormal bone remodelling within the otic capsule, leading to formation of sclerotic lesions of the temporal bone. Encroachment of these lesions on to the footplate of the stapes in the middle ear leads to stapes fixation and subsequent conductive hearing loss. The hereditary nature of otosclerosis has long been recognised due to its recurrence within families, but its genetic aetiology is yet to be characterised. Although many familial linkage studies and candidate gene association studies to investigate the genetic nature of otosclerosis have been performed in recent years, progress in identifying disease causing genes has been slow. This is largely due to the highly heterogeneous nature of this condition. The research presented in this thesis examines the molecular and genetic basis of otosclerosis using two next generation sequencing technologies; RNA-sequencing and Whole Exome Sequencing. RNA–sequencing has provided human stapes transcriptomes for healthy and diseased stapes, and in combination with pathway analysis has helped identify genes and molecular processes dysregulated in otosclerotic tissue. Whole Exome Sequencing has been employed to investigate rare variants that segregate with otosclerosis in affected families, and has been followed by a variant filtering strategy, which has prioritised genes found to be dysregulated during RNA-sequencing. -

Indian Journal for the Practicing Doctor Vol 7 Issue 1
ISSN : 0973-516X IJPD Vol. 7 Issue 1 Vol. 7, Issue 1, Jan- Feb 2012 IInnddiiaann JJoouurrnnaall ffoorr TThhee PPrraaccttiicciinngg DDooccttoorr Editor-in-Chief Saleem-ur-Rehman Executive Editor SM Kadri Electronic Version (full text) www.ijpd.pbworks.com 2 IJPD Vol. 7 Issue 1 INDIAN JOURNAL FOR THE PRACTISING DOCTOR IJPD (An Official Publication of Directorate of Health Services, Kashmir) www.ijpd.pbworks.com Editor in Chief Saleem-ur-Rehman Director Health Services, Kashmir, JK Executive Editor SM Kadri Epidemiologist, Kashmir Province, Directorate of Health Services, Kashmir, JK Assistant Editors Chinmay Shah Associate Professor, Department of Physiology Government Medical College Bhavnagar, Gujarat. India Rehana Kounsar State Surveillance Officer, IDSP/NCD Directorate of Health Services, Kashmir, JK Jehangir Bakshi Member Secretary, K-RICH (Kashmir Reforms and Innovations Committee for Healthcare) Directorate of Health Services, Kashmir, JK National Advisory Board Muzaffar Ahmed Anmol Gupta Member National Disaster Management Professor, Department of Community Medicine, Authority,New Delhi, India IGMC, Shimla, India Showkat Zargar Harish Pemde Director, Sheri- Kashmir Institute of Medical Science Professor of Pediatrics, Lady Hardinge Medical (SKIMS) , Srinagar , JK College, Kalawati Saran Children's Hospital, Delhi. Qazi Masood Ahmed D. Thamma Rao Principal, Government Medical College Senior Advisor, Public Health Foundation of India Srinagar, JK New Delhi Abdul Hamid Zargar Yogeshwar Gupta Member Institute Body, AIIMS, Specialist in Hospital and Health Care Management, Chairman, Independent Ethics Committee JK Fortis- Escorts Hospital & Research Centre, New Delhi Kanav Kahol Parvez Koul Team Leader, Division of Affordable Head, Department of Medicine Health Technologies, PHFI, Sheri- Kashmir Institute of Medical Science New Delhi, India (SKIMS) Srinagar, JK Tarun Seem Masood Tanvir IRS, Addl. -

1 Supporting Information for a Microrna Network Regulates
Supporting Information for A microRNA Network Regulates Expression and Biosynthesis of CFTR and CFTR-ΔF508 Shyam Ramachandrana,b, Philip H. Karpc, Peng Jiangc, Lynda S. Ostedgaardc, Amy E. Walza, John T. Fishere, Shaf Keshavjeeh, Kim A. Lennoxi, Ashley M. Jacobii, Scott D. Rosei, Mark A. Behlkei, Michael J. Welshb,c,d,g, Yi Xingb,c,f, Paul B. McCray Jr.a,b,c Author Affiliations: Department of Pediatricsa, Interdisciplinary Program in Geneticsb, Departments of Internal Medicinec, Molecular Physiology and Biophysicsd, Anatomy and Cell Biologye, Biomedical Engineeringf, Howard Hughes Medical Instituteg, Carver College of Medicine, University of Iowa, Iowa City, IA-52242 Division of Thoracic Surgeryh, Toronto General Hospital, University Health Network, University of Toronto, Toronto, Canada-M5G 2C4 Integrated DNA Technologiesi, Coralville, IA-52241 To whom correspondence should be addressed: Email: [email protected] (M.J.W.); yi- [email protected] (Y.X.); Email: [email protected] (P.B.M.) This PDF file includes: Materials and Methods References Fig. S1. miR-138 regulates SIN3A in a dose-dependent and site-specific manner. Fig. S2. miR-138 regulates endogenous SIN3A protein expression. Fig. S3. miR-138 regulates endogenous CFTR protein expression in Calu-3 cells. Fig. S4. miR-138 regulates endogenous CFTR protein expression in primary human airway epithelia. Fig. S5. miR-138 regulates CFTR expression in HeLa cells. Fig. S6. miR-138 regulates CFTR expression in HEK293T cells. Fig. S7. HeLa cells exhibit CFTR channel activity. Fig. S8. miR-138 improves CFTR processing. Fig. S9. miR-138 improves CFTR-ΔF508 processing. Fig. S10. SIN3A inhibition yields partial rescue of Cl- transport in CF epithelia. -

Structural Basis for DEAH-Helicase Activation by G-Patch Proteins
Structural basis for DEAH-helicase activation by G-patch proteins Michael K. Studera, Lazar Ivanovica, Marco E. Webera, Sabrina Martia, and Stefanie Jonasa,1 aInstitute of Molecular Biology and Biophysics, Department of Biology, Swiss Federal Institute of Technology (ETH) Zürich, 8093 Zürich, Switzerland Edited by Joseph D. Puglisi, Stanford University School of Medicine, Stanford, CA, and approved February 21, 2020 (received for review August 12, 2019) RNA helicases of the DEAH/RHA family are involved in many essential RNA bases are stacked in the RNA binding channel between a long cellular processes, such as splicing or ribosome biogenesis, where β-hairpin in RecA2 (β14 to β15 in hsDHX15/scPrp43; SI Appendix, they remodel large RNA–protein complexes to facilitate transitions Fig. S1) and a conserved loop in RecA1 (termed “Hook-turn”). to the next intermediate. DEAH helicases couple adenosine tri- This means that during progression into the open state, the phosphate (ATP) hydrolysis to conformational changes of their β-hairpin and two other RNA-binding patches in RecA2 (termed catalytic core. This movement results in translocation along RNA, “Hook-loop” and “motif V”; SI Appendix, Fig. S1) have to shift 1 which is held in place by auxiliary C-terminal domains. The activity nucleotide (nt) toward the 5′ end of the RNA. Thus, when the of DEAH proteins is strongly enhanced by the large and diverse RecA domains close back up, at the start of the next hydrolysis class of G-patch activators. Despite their central roles in RNA me- cycle, the RNA is pushed by 1 nt through the RNA channel. -

The Function and Evolution of C2H2 Zinc Finger Proteins and Transposons
The function and evolution of C2H2 zinc finger proteins and transposons by Laura Francesca Campitelli A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Department of Molecular Genetics University of Toronto © Copyright by Laura Francesca Campitelli 2020 The function and evolution of C2H2 zinc finger proteins and transposons Laura Francesca Campitelli Doctor of Philosophy Department of Molecular Genetics University of Toronto 2020 Abstract Transcription factors (TFs) confer specificity to transcriptional regulation by binding specific DNA sequences and ultimately affecting the ability of RNA polymerase to transcribe a locus. The C2H2 zinc finger proteins (C2H2 ZFPs) are a TF class with the unique ability to diversify their DNA-binding specificities in a short evolutionary time. C2H2 ZFPs comprise the largest class of TFs in Mammalian genomes, including nearly half of all Human TFs (747/1,639). Positive selection on the DNA-binding specificities of C2H2 ZFPs is explained by an evolutionary arms race with endogenous retroelements (EREs; copy-and-paste transposable elements), where the C2H2 ZFPs containing a KRAB repressor domain (KZFPs; 344/747 Human C2H2 ZFPs) are thought to diversify to bind new EREs and repress deleterious transposition events. However, evidence of the gain and loss of KZFP binding sites on the ERE sequence is sparse due to poor resolution of ERE sequence evolution, despite the recent publication of binding preferences for 242/344 Human KZFPs. The goal of my doctoral work has been to characterize the Human C2H2 ZFPs, with specific interest in their evolutionary history, functional diversity, and coevolution with LINE EREs. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -
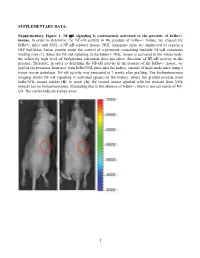
Activation of NF-Κb Signaling Promotes Prostate Cancer Progression in the Mouse and Predicts Poor Progression and Death in Pati
SUPPLEMENTARY DATA: Supplementary Figure 1. NF-B signaling is continuously activated in the prostate of - mouse. In order to determine the NF- '- mouse, we crossed the '- mice with NGL, a NF-B reporter mouse. NGL transgenic mice are engineered to express a GFP/luciferase fusion protein under the control of a promoter containing multiple NF-B consensus binding sites (1). Since the NF-'- NGL mouse is activated in the whole body, the relatively high level of background activation does not allow detection of NF- prostate. Therefore, in order to determine the NF-the '- mouse, we grafted the prostates from 'o the kidney capsule of male nude mice using a tissue rescue technique. NF-B activity was measured at 7 weeks after grafting. The bioluminescence imaging shows NF-B signaling is activated (green) in the kidney, where the grafted prostate from ' use resides (B). In panel (A), the control mouse (grafted with the prostate from NGL mouse) has no bioluminescence, illustrating that in the absence of '-, there is not activation of NF- B. The circles indicate kidney areas. 1 Supplementary Figure 2. NF-B signaling activated in the prostate of Myc/IB bigenic mouse. The prostates from Myc alone (Myc) and bigenic (Myc/IB) mice were harvested at 6 months of age. Activation of NF-B signaling in the prostate was determined by IHC staining of p65-pho antibody. 2 Supplementary Figure 3. Continuous activation of NF-B signaling promotes PCa progression in the Hi-Myc transgenic mouse. The prostates from Myc alone (Myc) and bigeneic (Myc/IB) mice were harvested at 6 months of age. -

DHX16 (NM 003587) Human Recombinant Protein Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TP302912 DHX16 (NM_003587) Human Recombinant Protein Product data: Product Type: Recombinant Proteins Description: Recombinant protein of human DEAH (Asp-Glu-Ala-His) box polypeptide 16 (DHX16) Species: Human Expression Host: HEK293T Tag: C-Myc/DDK Predicted MW: 119.1 kDa Concentration: >50 ug/mL as determined by microplate BCA method Purity: > 80% as determined by SDS-PAGE and Coomassie blue staining Buffer: 25 mM Tris.HCl, pH 7.3, 100 mM glycine, 10% glycerol Preparation: Recombinant protein was captured through anti-DDK affinity column followed by conventional chromatography steps. Storage: Store at -80°C. Stability: Stable for 12 months from the date of receipt of the product under proper storage and handling conditions. Avoid repeated freeze-thaw cycles. RefSeq: NP_003578 Locus ID: 8449 UniProt ID: O60231, Q5SQH4 RefSeq Size: 3477 Cytogenetics: 6p21.33 RefSeq ORF: 3126 Synonyms: DBP2; DDX16; NMOAS; PRO2014; Prp2; PRP8; PRPF2 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 DHX16 (NM_003587) Human Recombinant Protein – TP302912 Summary: DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. -

Whole Exome Sequencing Identifies Causative Mutations in the Majority of Consanguineous Or Familial Cases with Childhood-Onset I
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Kidney Manuscript Author Int. Author manuscript; Manuscript Author available in PMC 2016 August 01. Published in final edited form as: Kidney Int. 2016 February ; 89(2): 468–475. doi:10.1038/ki.2015.317. Whole exome sequencing identifies causative mutations in the majority of consanguineous or familial cases with childhood- onset increased renal echogenicity Daniela A. Braun#1, Markus Schueler#1, Jan Halbritter1, Heon Yung Gee1, Jonathan D. Porath1, Jennifer A. Lawson1, Rannar Airik1, Shirlee Shril1, Susan J. Allen2, Deborah Stein1, Adila Al Kindy3, Bodo B. Beck4, Nurcan Cengiz5, Khemchand N. Moorani6, Fatih Ozaltin7,8,9, Seema Hashmi10, John A. Sayer11, Detlef Bockenhauer12, Neveen A. Soliman13,14, Edgar A. Otto2, Richard P. Lifton15,16,17, and Friedhelm Hildebrandt1,17 1Division of Nephrology, Department of Medicine, Boston Children's Hospital, Harvard Medical School, Boston, Massachusetts, USA 2Department of Pediatrics, University of Michigan, Michigan, USA 3Department of Genetics, Sultan Qaboos University Hospital, Sultanate of Oman 4Institute for Human Genetics, University of Cologne, Germany 5Baskent University, School of Medicine, Adana Medical Training and Research Center, Department of Pediatric Nephrology, Adana, Turkey 6Department of Pediatric Nephrology, National Institute of Child Health, Karachi 75510, Pakistan 7Faculty of Medicine, Department of Pediatric Nephrology, Hacettepe University, Ankara, Turkey 8Nephrogenetics Laboratory, Faculty of Medicine, -

UC San Francisco Previously Published Works
UCSF UC San Francisco Previously Published Works Title FitSNPs: highly differentially expressed genes are more likely to have variants associated with disease. Permalink https://escholarship.org/uc/item/91k8p2km Journal Genome biology, 9(12) ISSN 1474-7596 Authors Chen, Rong Morgan, Alex A Dudley, Joel et al. Publication Date 2008 DOI 10.1186/gb-2008-9-12-r170 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Open Access Research2008ChenetVolume al. 9, Issue 12, Article R170 FitSNPs: highly differentially expressed genes are more likely to have variants associated with disease Rong Chen*†‡, Alex A Morgan*†‡, Joel Dudley*†‡, Tarangini Deshpande§, Li Li†, Keiichi Kodama*†‡, Annie P Chiang*†‡ and Atul J Butte*†‡ Addresses: *Stanford Center for Biomedical Informatics Research, 251 Cmpus Drive, Stanford, CA 94305, USA. †Department of Pediatrics, Stanford University School of Medicine, Stanford, CA 94305, USA. ‡Lucile Packard Children's Hospital, 725 Welch Road, Palo Alto, CA 94304, USA. §NuMedii Inc., Menlo Park, CA 94025, USA. Correspondence: Atul J Butte. Email: [email protected] Published: 5 December 2008 Received: 17 June 2008 Revised: 26 September 2008 Genome Biology 2008, 9:R170 (doi:10.1186/gb-2008-9-12-r170) Accepted: 5 December 2008 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2008/9/12/R170 © 2008 Chen et al.; licensee BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. -

Differential Expression of Skin Cancer and Hair-Follicle Cycle Regulated Genes in Tumor Susceptible K14-Agouti Mice
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 8-2006 Differential Expression of Skin Cancer and Hair-Follicle Cycle Regulated Genes in Tumor Susceptible K14-Agouti Mice Yesim Aydin Son University of Tennessee - Knoxville Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Part of the Life Sciences Commons Recommended Citation Son, Yesim Aydin, "Differential Expression of Skin Cancer and Hair-Follicle Cycle Regulated Genes in Tumor Susceptible K14-Agouti Mice. " PhD diss., University of Tennessee, 2006. https://trace.tennessee.edu/utk_graddiss/1636 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by Yesim Aydin Son entitled "Differential Expression of Skin Cancer and Hair-Follicle Cycle Regulated Genes in Tumor Susceptible K14-Agouti Mice." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the equirr ements for the degree of Doctor of Philosophy, with a major in Life Sciences. Edward J. Michaud, Major Professor We have read this dissertation and recommend its acceptance: Cymbeline T. Culiat, Mitchel -

Mrna Editing, Processing and Quality Control in Caenorhabditis Elegans
| WORMBOOK mRNA Editing, Processing and Quality Control in Caenorhabditis elegans Joshua A. Arribere,*,1 Hidehito Kuroyanagi,†,1 and Heather A. Hundley‡,1 *Department of MCD Biology, UC Santa Cruz, California 95064, †Laboratory of Gene Expression, Medical Research Institute, Tokyo Medical and Dental University, Tokyo 113-8510, Japan, and ‡Medical Sciences Program, Indiana University School of Medicine-Bloomington, Indiana 47405 ABSTRACT While DNA serves as the blueprint of life, the distinct functions of each cell are determined by the dynamic expression of genes from the static genome. The amount and specific sequences of RNAs expressed in a given cell involves a number of regulated processes including RNA synthesis (transcription), processing, splicing, modification, polyadenylation, stability, translation, and degradation. As errors during mRNA production can create gene products that are deleterious to the organism, quality control mechanisms exist to survey and remove errors in mRNA expression and processing. Here, we will provide an overview of mRNA processing and quality control mechanisms that occur in Caenorhabditis elegans, with a focus on those that occur on protein-coding genes after transcription initiation. In addition, we will describe the genetic and technical approaches that have allowed studies in C. elegans to reveal important mechanistic insight into these processes. KEYWORDS Caenorhabditis elegans; splicing; RNA editing; RNA modification; polyadenylation; quality control; WormBook TABLE OF CONTENTS Abstract 531 RNA Editing and Modification 533 Adenosine-to-inosine RNA editing 533 The C. elegans A-to-I editing machinery 534 RNA editing in space and time 535 ADARs regulate the levels and fates of endogenous dsRNA 537 Are other modifications present in C.