A Molecular and Genetic Analysis of Otosclerosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
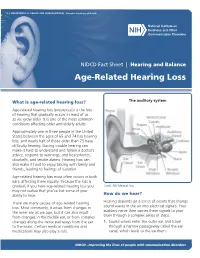
Age-Related Hearing Loss
U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES ∙ National Institutes of Health NIDCD Fact Sheet | Hearing and Balance Age-Related Hearing Loss What is age-related hearing loss? The auditory system Age-related hearing loss (presbycusis) is the loss of hearing that gradually occurs in most of us as we grow older. It is one of the most common conditions affecting older and elderly adults. Approximately one in three people in the United States between the ages of 65 and 74 has hearing loss, and nearly half of those older than 75 have difficulty hearing. Having trouble hearing can make it hard to understand and follow a doctor’s advice, respond to warnings, and hear phones, doorbells, and smoke alarms. Hearing loss can also make it hard to enjoy talking with family and friends, leading to feelings of isolation. Age-related hearing loss most often occurs in both ears, affecting them equally. Because the loss is gradual, if you have age-related hearing loss you Credit: NIH Medical Arts may not realize that you’ve lost some of your ability to hear. How do we hear? There are many causes of age-related hearing Hearing depends on a series of events that change loss. Most commonly, it arises from changes in sound waves in the air into electrical signals. Your the inner ear as we age, but it can also result auditory nerve then carries these signals to your from changes in the middle ear, or from complex brain through a complex series of steps. changes along the nerve pathways from the ear 1. -

Novel Human Embryonic Stem Cell Regulators Identified by Conserved and Distinct Cpg Island Methylation State
RESEARCH ARTICLE Novel Human Embryonic Stem Cell Regulators Identified by Conserved and Distinct CpG Island Methylation State Steve Pells1,2*, Eirini Koutsouraki1, Sofia Morfopoulou1, Sara Valencia-Cadavid1, Simon R. Tomlinson1, Ravi Kalathur3, Matthias E. Futschik3, Paul A. De Sousa1,2* 1 MRC Centre for Regenerative Medicine, School of Clinical Studies, University of Edinburgh, Edinburgh, EH16 4SB, United Kingdom, 2 Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, EH16 4SB, United Kingdom, 3 Centre for Molecular and Structural Biomedicine, University of Algarve, 8005–139, Faro, Portugal * [email protected] (PDS); [email protected] (SP) Abstract Human embryonic stem cells (hESCs) undergo epigenetic changes in vitro which may com- OPEN ACCESS promise function, so an epigenetic pluripotency “signature” would be invaluable for line vali- Citation: Pells S, Koutsouraki E, Morfopoulou S, dation. We assessed Cytosine-phosphate-Guanine Island (CGI) methylation in hESCs by Valencia-Cadavid S, Tomlinson SR, Kalathur R, et al. genomic DNA hybridisation to a CGI array, and saw substantial variation in CGI methylation (2015) Novel Human Embryonic Stem Cell Regulators Identified by Conserved and Distinct CpG between lines. Comparison of hESC CGI methylation profiles to corresponding somatic Island Methylation State. PLoS ONE 10(7): tissue data and hESC mRNA expression profiles identified a conserved hESC-specific e0131102. doi:10.1371/journal.pone.0131102 methylation pattern associated with expressed genes. Transcriptional repressors and acti- Editor: Jason Glenn Knott, Michigan State University, vators were over-represented amongst genes whose associated CGIs were methylated or UNITED STATES unmethylated specifically in hESCs, respectively. Knockdown of candidate transcriptional Received: March 10, 2015 regulators (HMGA1, GLIS2, PFDN5) induced differentiation in hESCs, whereas ectopic Accepted: May 27, 2015 expression in fibroblasts modulated iPSC colony formation. -

PLZF Targets Developmental Enhancers for Activation During Osteogenic Differentiation of Human Mesenchymal Stem Cells
RESEARCH ARTICLE PLZF targets developmental enhancers for activation during osteogenic differentiation of human mesenchymal stem cells Shuchi Agrawal Singh1,2,3*, Mads Lerdrup1,3, Ana-Luisa R Gomes1,3, Harmen JG van de Werken4,5,6, Jens Vilstrup Johansen1,3,7, Robin Andersson1,3,7, Albin Sandelin1,3,7, Kristian Helin8,9,10, Klaus Hansen1,3* 1Biotech Research and Innovation Centre (BRIC), Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark; 2Department of Hematology, Cambridge Institute for Medical Research and Welcome Trust/MRC Stem Cell Institute, University of Cambridge, Cambridge, United Kingdom; 3Centre for Epigenetics, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark; 4Department of Cell Biology, University Medical Center, Rotterdam, Netherlands; 5Cancer Computational Biology Center, University Medical Center, Rotterdam, Netherlands; 6Department of Urology, University Medical Center, Rotterdam, Netherlands; 7Department of Biology, The Bioinformatics Centre, University of Copenhagen, Copenhagen, Denmark; 8The Novo Nordisk Center for Stem Cell Biology, Faculty of Health and Medical Sciences University of Copenhagen, Copenhagen, Denmark; 9Cell Biology Program, Memorial Sloan Kettering Cancer Center, New York, United States; 10Center for Epigenetics Research, Memorial Sloan Kettering Cancer Center, New York, United States *For correspondence: [email protected]; Abstract The PLZF transcription factor is essential for osteogenic differentiation of hMSCs; [email protected] (SAS); however, its regulation and molecular function during this process is not fully understood. Here, we [email protected] (KHA) revealed that the ZBTB16 locus encoding PLZF, is repressed by Polycomb (PcG) and H3K27me3 in Competing interests: The naive hMSCs. At the pre-osteoblast stage of differentiation, the locus lost PcG binding and authors declare that no H3K27me3, gained JMJD3 recruitment, and H3K27ac resulting in high expression of PLZF. -

Low Basal Expression and Slow Induction of IFITM3 Puts Immune
bioRxiv preprint doi: https://doi.org/10.1101/2019.12.20.885590; this version posted December 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 TITLE: Low basal expression and slow induction of IFITM3 puts immune 2 cells at risk of influenza A infection 3 4 Running head (40 characters): 5 Cells at risk of Influenza A infection 6 7 Authors: 8 Dannielle Wellington1,2,*, , Zixi Yin1,2, Liwei Zhang1, Jessica Forbester1,3, Kerry 9 Kite1, Henry Laurenson-Schafer1, Shokouh Makvandi-Nejad1 , Boquan Jin4, 10 Emma Bowes5, Krishnageetha Manoharan5, David Maldonado-Perez5, Clare 11 Verill5, Ian Humphreys3 & Tao Dong1,2,* 12 1. MRC Human Immunology Unit, MRC Weatherall Institute of Molecular Medicine, Radcliffe 13 Department of Medicine, Oxford University, Oxford OX3 9DS, UK 14 2. Chinese Academy of Medical Sciences (CAMS) Oxford Institute, Nuffield Department of 15 Medicine, Oxford University, OX3 7BN, UK 16 3. Division of Infection and Immunity/Systems Immunity University Research Institute, Cardiff 17 University, Cardiff, CF14 4XN, UK 18 4. Fourth Military Medical University, Xian, China 19 5. Oxford Radcliffe BioBank, Nuffield Department of Surgical Sciences, Oxford University, 20 Oxford OX3 9DU, UK 21 *Corresponding to Dannielle Wellington [email protected] or Tao Dong, 22 [email protected] 23 24 Acknowledgements: 25 The authors would like to thank the following facilities and individuals within 26 the MRC Weatherall Institute (Oxford) for their invaluaBle help in the following 27 experiments: Mass Cytometry Facility and Alain Townsend for kindly providing 28 virus stocks. -

V10a91-Tasheva Pgmkr
Molecular Vision 2004; 10:758-772 <http://www.molvis.org/molvis/v10/a91> ©2004 Molecular Vision Received 12 August 2004 | Accepted 7 October 2004 | Published 7 October 2004 Analysis of transcriptional regulation of the small leucine rich proteoglycans Elena S. Tasheva,1 Bernward Klocke,2 Gary W. Conrad1 1Kansas State University, Division of Biology, Manhattan, KS; 2Genomatix Software GmbH, München, Germany Purpose: Small leucine rich proteoglycans (SLRPs) constitute a family of secreted proteoglycans that are important for collagen fibrillogenesis, cellular growth, differentiation, and migration. Ten of the 13 known members of the SLRP gene family are arranged in tandem clusters on human chromosomes 1, 9, and 12. Their syntenic equivalents are on mouse chromosomes 1, 13, and 10, and rat chromosomes 13, 17, and 7. The purpose of this study was to determine whether there is evidence for control elements, which could regulate the expression of these clusters coordinately. Methods: Promoters were identified using a comparative genomics approach and Genomatix software tools. For each gene a set of human, mouse, and rat orthologous promoters was extracted from genomic sequences. Transcription factor (TF) binding site analysis combined with a literature search was performed using MatInspector and Genomatix’ BiblioSphere. Inspection for the presence of interspecies conserved scaffold/matrix attachment regions (S/MARs) was performed using ElDorado annotation lists. DNAseI hypersensitivity assay, chromatin immunoprecipitation (ChIP), and transient transfection experiments were used to validate the results from bioinformatics analysis. Results: Transcription factor binding site analysis combined with a literature search revealed co-citations between several SLRPs and TFs Runx2 and IRF1, indicating that these TFs have potential roles in transcriptional regulation of the SLRP family members. -

PRESBYCUSIS Diagnosis and Treatment
Hear the FACTS about PRESBYCUSIS Diagnosis and Treatment NORMAL HEARING What is Presbycusis? FREQUENCY (in Hertz) . A gradual reduction in hearing as we get older, typically affecting both 250 500 1000 2000 4000 8000 -10 ears equally. 0 X 10 X X Common in men and women, with men typically having greater X •X . 20 • •X • • • 30 hearing loss than women of the same age. 40 Typically a greater hearing loss for high frequency sounds than for low 50 . (in dBHL) 60 INTENSITY frequency sounds. 70 80 A treatable condition that can benefit greatly from technological 90 . advances in various amplification or hearing assistance devices, along 100 • Right Ear 110 X Left Ear with counseling on effective communication strategies. PRESBYCUSIS HEARING LOSS What causes Presbycusis? FREQUENCY (in Hertz) . Family history or hereditary factors. 250 500 1000 2000 4000 8000 -10 Changes in the inner ear blood supply related to heart disease, 0 . 10 •X diabetes, high blood pressure and smoking. 20 •X 30 •X A loss of sound sensitivity from cumulative exposure to loud sounds. 40 •X . 50 (in dBHL) 60 INTENSITY X 70 • Symptoms: X 80 • •X 90 . Frequently asking people to repeat what they say, especially in 100 Right Ear “difficult listening places”. 110 X Left Ear . Ability to hear lower-pitched men's voices easier than higher-pitched NORMAL INNER EAR PRESBYCUSIC INNER EAR women’s or children’s voices. People complaining that the TV is being played too loud. Tinnitus, also known as “head noise”, which produces buzzing or ringing sounds in the ear. Diagnosis: . Talk honestly with your Hearing Healthcare Provider about daily hearing problems. -

Mir-96 Regulates the Progression of Differentiation in Mammalian Cochlear Inner and Outer Hair Cells
miR-96 regulates the progression of differentiation in mammalian cochlear inner and outer hair cells Stephanie Kuhna,1, Stuart L. Johnsona,1, David N. Furnessb, Jing Chenc, Neil Inghamc, Jennifer M. Hiltonc, Georg Steffesc, Morag A. Lewisc, Valeria Zampinia,d, Carole M. Hackneya, Sergio Masettod, Matthew C. Holleya, Karen P. Steelc, and Walter Marcottia,2 aDepartment of Biomedical Science, University of Sheffield, Sheffield S10 2TN, United Kingdom; bInstitute for Science and Technology in Medicine, Keele University, Keele ST5 5BG, United Kingdom; cWellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, United Kingdom; and dDepartment of Physiology, University of Pavia, 27100 Pavia, Italy Edited by Mary-Claire King, University of Washington, Seattle, WA, and approved December 27, 2010 (received for review November 8, 2010) MicroRNAs (miRNAs) are small noncoding RNAs able to regulate cells up to at least P5 (23) and in the spiral ganglion up to P14 a broad range of protein-coding genes involved in many biological (19). Mutations in miR-96 have been associated with non- processes. miR-96 is a sensory organ-specific miRNA expressed in syndromic progressive hearing loss in humans (15) and mice (23). the mammalian cochlea during development. Mutations in miR-96 We found that hair cell development in diminuendo mice with cause nonsyndromic progressive hearing loss in humans and mice. a Mir96 mutation is arrested at around the day of birth, a time The mouse mutant diminuendo has a single base change in the when IHCs and OHCs still exhibit qualitatively similar bio- seed region of the Mir96 gene leading to widespread changes in physical properties. -

Characterizing G-Quadruplex Mediated Regulation of the Amyloid Precursor Protein Expression by Ezekiel Matthew David Crenshaw Oc
I Characterizing G-Quadruplex Mediated Regulation of the Amyloid Precursor Protein Expression By Ezekiel Matthew David Crenshaw October, 2016 A Dissertation Presented to the Faculty of Drexel University Department of Biology In partial fulfillment of the Requirements for the Degree of Ph.D. Daniel Marenda, Ph.D. (Chair) Elias Spiliotis, Ph.D. Associate Professor, Associate Professor, Director, Graduate Student Program Director, Cell Imaging Center Co-Director, Cell Imaging Center Dept. Biology Dept. Biology Drexel University Drexel University Tali Gidalevitz, Ph.D. Aleister Saunders, Ph.D. (Advisor) Assistant Professor, Dept. Biology Associate Professor, Drexel University Director of the RNAi Resource Center, Dept. Biology Jeff Twiss, Ph.D. Senior Vice Provost for Research, Drexel University Professor, SmartState Chair, Childhood Neurotherapeutics Michael Akins, Ph.D.(Co-Advisor) Dept. Biology University of South Carolina Assistant Professor, Dept. Biology Drexel University II Acknowledgements Proverbs 3:5-6, “Trust in the Lord with all thine heart and lean not unto thine own understanding. In all thine ways acknowledge Him and He shall direct thy paths” I want to start off my acknowledging my LORD, who is known by many names. He is El Elyon, The Most High God; He is YAHWEH Shammah, for He has been there for me; He is YAHWEH Yireh, for He has provided for all of my needs; He is YAHWEH Shalom, for He is my peace. He is Yeshua Ha Mashiach, for He is my Savior. Coming from humble beginnings, my faith in the LORD has been the source of strength and encouragement to overcome the many obstacles I faced growing up, as well as in my pursuit for my Ph.D. -

Nucleolin and Its Role in Ribosomal Biogenesis
NUCLEOLIN: A NUCLEOLAR RNA-BINDING PROTEIN INVOLVED IN RIBOSOME BIOGENESIS Inaugural-Dissertation zur Erlangung des Doktorgrades der Mathematisch-Naturwissenschaftlichen Fakultät der Heinrich-Heine-Universität Düsseldorf vorgelegt von Julia Fremerey aus Hamburg Düsseldorf, April 2016 2 Gedruckt mit der Genehmigung der Mathematisch-Naturwissenschaftlichen Fakultät der Heinrich-Heine-Universität Düsseldorf Referent: Prof. Dr. A. Borkhardt Korreferent: Prof. Dr. H. Schwender Tag der mündlichen Prüfung: 20.07.2016 3 Die vorgelegte Arbeit wurde von Juli 2012 bis März 2016 in der Klinik für Kinder- Onkologie, -Hämatologie und Klinische Immunologie des Universitätsklinikums Düsseldorf unter Anleitung von Prof. Dr. A. Borkhardt und in Kooperation mit dem ‚Laboratory of RNA Molecular Biology‘ an der Rockefeller Universität unter Anleitung von Prof. Dr. T. Tuschl angefertigt. 4 Dedicated to my family TABLE OF CONTENTS 5 TABLE OF CONTENTS TABLE OF CONTENTS ............................................................................................... 5 LIST OF FIGURES ......................................................................................................10 LIST OF TABLES .......................................................................................................12 ABBREVIATION .........................................................................................................13 ABSTRACT ................................................................................................................19 ZUSAMMENFASSUNG -

Genome-Wide DNA Methylation Analysis of KRAS Mutant Cell Lines Ben Yi Tew1,5, Joel K
www.nature.com/scientificreports OPEN Genome-wide DNA methylation analysis of KRAS mutant cell lines Ben Yi Tew1,5, Joel K. Durand2,5, Kirsten L. Bryant2, Tikvah K. Hayes2, Sen Peng3, Nhan L. Tran4, Gerald C. Gooden1, David N. Buckley1, Channing J. Der2, Albert S. Baldwin2 ✉ & Bodour Salhia1 ✉ Oncogenic RAS mutations are associated with DNA methylation changes that alter gene expression to drive cancer. Recent studies suggest that DNA methylation changes may be stochastic in nature, while other groups propose distinct signaling pathways responsible for aberrant methylation. Better understanding of DNA methylation events associated with oncogenic KRAS expression could enhance therapeutic approaches. Here we analyzed the basal CpG methylation of 11 KRAS-mutant and dependent pancreatic cancer cell lines and observed strikingly similar methylation patterns. KRAS knockdown resulted in unique methylation changes with limited overlap between each cell line. In KRAS-mutant Pa16C pancreatic cancer cells, while KRAS knockdown resulted in over 8,000 diferentially methylated (DM) CpGs, treatment with the ERK1/2-selective inhibitor SCH772984 showed less than 40 DM CpGs, suggesting that ERK is not a broadly active driver of KRAS-associated DNA methylation. KRAS G12V overexpression in an isogenic lung model reveals >50,600 DM CpGs compared to non-transformed controls. In lung and pancreatic cells, gene ontology analyses of DM promoters show an enrichment for genes involved in diferentiation and development. Taken all together, KRAS-mediated DNA methylation are stochastic and independent of canonical downstream efector signaling. These epigenetically altered genes associated with KRAS expression could represent potential therapeutic targets in KRAS-driven cancer. Activating KRAS mutations can be found in nearly 25 percent of all cancers1. -

S41467-020-18249-3.Pdf
ARTICLE https://doi.org/10.1038/s41467-020-18249-3 OPEN Pharmacologically reversible zonation-dependent endothelial cell transcriptomic changes with neurodegenerative disease associations in the aged brain Lei Zhao1,2,17, Zhongqi Li 1,2,17, Joaquim S. L. Vong2,3,17, Xinyi Chen1,2, Hei-Ming Lai1,2,4,5,6, Leo Y. C. Yan1,2, Junzhe Huang1,2, Samuel K. H. Sy1,2,7, Xiaoyu Tian 8, Yu Huang 8, Ho Yin Edwin Chan5,9, Hon-Cheong So6,8, ✉ ✉ Wai-Lung Ng 10, Yamei Tang11, Wei-Jye Lin12,13, Vincent C. T. Mok1,5,6,14,15 &HoKo 1,2,4,5,6,8,14,16 1234567890():,; The molecular signatures of cells in the brain have been revealed in unprecedented detail, yet the ageing-associated genome-wide expression changes that may contribute to neurovas- cular dysfunction in neurodegenerative diseases remain elusive. Here, we report zonation- dependent transcriptomic changes in aged mouse brain endothelial cells (ECs), which pro- minently implicate altered immune/cytokine signaling in ECs of all vascular segments, and functional changes impacting the blood–brain barrier (BBB) and glucose/energy metabolism especially in capillary ECs (capECs). An overrepresentation of Alzheimer disease (AD) GWAS genes is evident among the human orthologs of the differentially expressed genes of aged capECs, while comparative analysis revealed a subset of concordantly downregulated, functionally important genes in human AD brains. Treatment with exenatide, a glucagon-like peptide-1 receptor agonist, strongly reverses aged mouse brain EC transcriptomic changes and BBB leakage, with associated attenuation of microglial priming. We thus revealed tran- scriptomic alterations underlying brain EC ageing that are complex yet pharmacologically reversible. -

Downloaded From
Antimicrobial Activity of PLUNC Protects against Pseudomonas aeruginosa Infection Lina Lukinskiene, Yang Liu, Susan D. Reynolds, Chad Steele, Barry R. Stripp, George D. Leikauf, Jay K. Kolls and This information is current as Y. Peter Di of October 2, 2021. J Immunol 2011; 187:382-390; Prepublished online 1 June 2011; doi: 10.4049/jimmunol.1001769 http://www.jimmunol.org/content/187/1/382 Downloaded from References This article cites 37 articles, 15 of which you can access for free at: http://www.jimmunol.org/content/187/1/382.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on October 2, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2011 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Antimicrobial Activity of PLUNC Protects against Pseudomonas aeruginosa Infection Lina Lukinskiene,* Yang Liu,* Susan D. Reynolds,† Chad Steele,‡ Barry R. Stripp,x George D. Leikauf,* Jay K. Kolls,{ and Y.