Microrna-1182 and Let-7A Exert Synergistic Inhibition on Invasion
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cytogenomic SNP Microarray - Fetal ARUP Test Code 2002366 Maternal Contamination Study Fetal Spec Fetal Cells
Patient Report |FINAL Client: Example Client ABC123 Patient: Patient, Example 123 Test Drive Salt Lake City, UT 84108 DOB 2/13/1987 UNITED STATES Gender: Female Patient Identifiers: 01234567890ABCD, 012345 Physician: Doctor, Example Visit Number (FIN): 01234567890ABCD Collection Date: 00/00/0000 00:00 Cytogenomic SNP Microarray - Fetal ARUP test code 2002366 Maternal Contamination Study Fetal Spec Fetal Cells Single fetal genotype present; no maternal cells present. Fetal and maternal samples were tested using STR markers to rule out maternal cell contamination. This result has been reviewed and approved by Maternal Specimen Yes Cytogenomic SNP Microarray - Fetal Abnormal * (Ref Interval: Normal) Test Performed: Cytogenomic SNP Microarray- Fetal (ARRAY FE) Specimen Type: Direct (uncultured) villi Indication for Testing: Patient with 46,XX,t(4;13)(p16.3;q12) (Quest: EN935475D) ----------------------------------------------------------------- ----- RESULT SUMMARY Abnormal Microarray Result (Male) Unbalanced Translocation Involving Chromosomes 4 and 13 Classification: Pathogenic 4p Terminal Deletion (Wolf-Hirschhorn syndrome) Copy number change: 4p16.3p16.2 loss Size: 5.1 Mb 13q Proximal Region Deletion Copy number change: 13q11q12.12 loss Size: 6.1 Mb ----------------------------------------------------------------- ----- RESULT DESCRIPTION This analysis showed a terminal deletion (1 copy present) involving chromosome 4 within 4p16.3p16.2 and a proximal interstitial deletion (1 copy present) involving chromosome 13 within 13q11q12.12. This -

A Molecular and Genetic Analysis of Otosclerosis
A molecular and genetic analysis of otosclerosis Joanna Lauren Ziff Submitted for the degree of PhD University College London January 2014 1 Declaration I, Joanna Ziff, confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. Where work has been conducted by other members of our laboratory, this has been indicated by an appropriate reference. 2 Abstract Otosclerosis is a common form of conductive hearing loss. It is characterised by abnormal bone remodelling within the otic capsule, leading to formation of sclerotic lesions of the temporal bone. Encroachment of these lesions on to the footplate of the stapes in the middle ear leads to stapes fixation and subsequent conductive hearing loss. The hereditary nature of otosclerosis has long been recognised due to its recurrence within families, but its genetic aetiology is yet to be characterised. Although many familial linkage studies and candidate gene association studies to investigate the genetic nature of otosclerosis have been performed in recent years, progress in identifying disease causing genes has been slow. This is largely due to the highly heterogeneous nature of this condition. The research presented in this thesis examines the molecular and genetic basis of otosclerosis using two next generation sequencing technologies; RNA-sequencing and Whole Exome Sequencing. RNA–sequencing has provided human stapes transcriptomes for healthy and diseased stapes, and in combination with pathway analysis has helped identify genes and molecular processes dysregulated in otosclerotic tissue. Whole Exome Sequencing has been employed to investigate rare variants that segregate with otosclerosis in affected families, and has been followed by a variant filtering strategy, which has prioritised genes found to be dysregulated during RNA-sequencing. -

Oncogenomics of C-Myc Transgenic Mice Reveal Novel Regulators of Extracellular Signaling, Angiogenesis and Invasion with Clinica
www.impactjournals.com/oncotarget/ Oncotarget, 2017, Vol. 8, (No. 60), pp: 101808-101831 Research Paper Oncogenomics of c-Myc transgenic mice reveal novel regulators of extracellular signaling, angiogenesis and invasion with clinical significance for human lung adenocarcinoma Yari Ciribilli1,2 and Jürgen Borlak2 1Centre for Integrative Biology (CIBIO), University of Trento, 38123 Povo (TN), Italy 2Centre for Pharmacology and Toxicology, Hannover Medical School, 30625 Hannover, Germany Correspondence to: Jürgen Borlak, email: [email protected] Keywords: c-Myc transgenic mouse model of lung cancer, papillary adenocarcinomas, whole genome scans, c-Myc regulatory gene networks, c-Myc targeted regulators of extracellular signaling Received: June 26, 2017 Accepted: September 21, 2017 Published: October 23, 2017 Copyright: Ciribilli et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT The c-Myc transcription factor is frequently deregulated in cancers. To search for disease diagnostic and druggable targets a transgenic lung cancer disease model was investigated. Oncogenomics identified c-Myc target genes in lung tumors. These were validated by RT-PCR, Western Blotting, EMSA assays and ChIP-seq data retrieved from public sources. Gene reporter and ChIP assays verified functional importance of c-Myc binding sites. The clinical significance was established by RT-qPCR in tumor and matched healthy control tissues, by RNA-seq data retrieved from the TCGA Consortium and by immunohistochemistry recovered from the Human Protein Atlas repository. In transgenic lung tumors 25 novel candidate genes were identified. -

Mir-96 Regulates the Progression of Differentiation in Mammalian Cochlear Inner and Outer Hair Cells
miR-96 regulates the progression of differentiation in mammalian cochlear inner and outer hair cells Stephanie Kuhna,1, Stuart L. Johnsona,1, David N. Furnessb, Jing Chenc, Neil Inghamc, Jennifer M. Hiltonc, Georg Steffesc, Morag A. Lewisc, Valeria Zampinia,d, Carole M. Hackneya, Sergio Masettod, Matthew C. Holleya, Karen P. Steelc, and Walter Marcottia,2 aDepartment of Biomedical Science, University of Sheffield, Sheffield S10 2TN, United Kingdom; bInstitute for Science and Technology in Medicine, Keele University, Keele ST5 5BG, United Kingdom; cWellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, United Kingdom; and dDepartment of Physiology, University of Pavia, 27100 Pavia, Italy Edited by Mary-Claire King, University of Washington, Seattle, WA, and approved December 27, 2010 (received for review November 8, 2010) MicroRNAs (miRNAs) are small noncoding RNAs able to regulate cells up to at least P5 (23) and in the spiral ganglion up to P14 a broad range of protein-coding genes involved in many biological (19). Mutations in miR-96 have been associated with non- processes. miR-96 is a sensory organ-specific miRNA expressed in syndromic progressive hearing loss in humans (15) and mice (23). the mammalian cochlea during development. Mutations in miR-96 We found that hair cell development in diminuendo mice with cause nonsyndromic progressive hearing loss in humans and mice. a Mir96 mutation is arrested at around the day of birth, a time The mouse mutant diminuendo has a single base change in the when IHCs and OHCs still exhibit qualitatively similar bio- seed region of the Mir96 gene leading to widespread changes in physical properties. -

SARS-Cov-2 Entry Protein TMPRSS2 and Its Homologue, TMPRSS4
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.26.441280; this version posted April 26, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 SARS-CoV-2 Entry Protein TMPRSS2 and Its 2 Homologue, TMPRSS4 Adopts Structural Fold Similar 3 to Blood Coagulation and Complement Pathway 4 Related Proteins ∗,a ∗∗,b b 5 Vijaykumar Yogesh Muley , Amit Singh , Karl Gruber , Alfredo ∗,a 6 Varela-Echavarría a 7 Instituto de Neurobiología, Universidad Nacional Autónoma de México, Querétaro, México b 8 Institute of Molecular Biosciences, University of Graz, Graz, Austria 9 Abstract The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) utilizes TMPRSS2 receptor to enter target human cells and subsequently causes coron- avirus disease 19 (COVID-19). TMPRSS2 belongs to the type II serine proteases of subfamily TMPRSS, which is characterized by the presence of the serine- protease domain. TMPRSS4 is another TMPRSS member, which has a domain architecture similar to TMPRSS2. TMPRSS2 and TMPRSS4 have been shown to be involved in SARS-CoV-2 infection. However, their normal physiological roles have not been explored in detail. In this study, we analyzed the amino acid sequences and predicted 3D structures of TMPRSS2 and TMPRSS4 to under- stand their functional aspects at the protein domain level. Our results suggest that these proteins are likely to have common functions based on their conserved domain organization. -

High-Throughput Characterization of Blood Serum Proteomics of IBD Patients with Respect to Aging and Genetic Factors
RESEARCH ARTICLE High-Throughput Characterization of Blood Serum Proteomics of IBD Patients with Respect to Aging and Genetic Factors Antonio F. Di Narzo1,2, Shannon E. Telesco3, Carrie Brodmerkel3, Carmen Argmann1,2, Lauren A. Peters2,4, Katherine Li3, Brian Kidd1,2, Joel Dudley1,2, Judy Cho1,2, Eric E. Schadt1,2, Andrew Kasarskis1,2, Radu Dobrin3*, Ke Hao1,2* 1 Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, United States of America, 2 Icahn Institute of Genomics and Multiscale Biology, Icahn School of a1111111111 Medicine at Mount Sinai, New York, New York, United States of America, 3 Janssen R&D, LLC, Spring a1111111111 House, Pennsylvania, United States of America, 4 Graduate School of Biomedical Sciences, Icahn School of a1111111111 Medicine at Mount Sinai, New York, New York, United States of America a1111111111 a1111111111 * [email protected] (RD); [email protected] (KH) Abstract OPEN ACCESS To date, no large scale, systematic description of the blood serum proteome has been per- Citation: Di Narzo AF, Telesco SE, Brodmerkel C, formed in inflammatory bowel disease (IBD) patients. By using microarray technology, a Argmann C, Peters LA, Li K, et al. (2017) High- more complete description of the blood proteome of IBD patients is feasible. It may help to Throughput Characterization of Blood Serum achieve a better understanding of the disease. We analyzed blood serum profiles of 1128 Proteomics of IBD Patients with Respect to Aging and Genetic Factors. PLoS Genet 13(1): e1006565. proteins in IBD patients of European descent (84 Crohn's Disease (CD) subjects and 88 doi:10.1371/journal.pgen.1006565 Ulcerative Colitis (UC) subjects) as well as 15 healthy control subjects, and linked protein Editor: Gregory S. -

Genome-Wide DNA Methylation Analysis Reveals Molecular Subtypes of Pancreatic Cancer
www.impactjournals.com/oncotarget/ Oncotarget, 2017, Vol. 8, (No. 17), pp: 28990-29012 Research Paper Genome-wide DNA methylation analysis reveals molecular subtypes of pancreatic cancer Nitish Kumar Mishra1 and Chittibabu Guda1,2,3,4 1Department of Genetics, Cell Biology and Anatomy, University of Nebraska Medical Center, Omaha, NE, 68198, USA 2Bioinformatics and Systems Biology Core, University of Nebraska Medical Center, Omaha, NE, 68198, USA 3Department of Biochemistry and Molecular Biology, University of Nebraska Medical Center, Omaha, NE, 68198, USA 4Fred and Pamela Buffet Cancer Center, University of Nebraska Medical Center, Omaha, NE, 68198, USA Correspondence to: Chittibabu Guda, email: [email protected] Keywords: TCGA, pancreatic cancer, differential methylation, integrative analysis, molecular subtypes Received: October 20, 2016 Accepted: February 12, 2017 Published: March 07, 2017 Copyright: Mishra et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Pancreatic cancer (PC) is the fourth leading cause of cancer deaths in the United States with a five-year patient survival rate of only 6%. Early detection and treatment of this disease is hampered due to lack of reliable diagnostic and prognostic markers. Recent studies have shown that dynamic changes in the global DNA methylation and gene expression patterns play key roles in the PC development; hence, provide valuable insights for better understanding the initiation and progression of PC. In the current study, we used DNA methylation, gene expression, copy number, mutational and clinical data from pancreatic patients. -

Mutations Suggests Both Loss-Of-Function and Gain-Of-Function Effects Morag A
© 2021. Published by The Company of Biologists Ltd | Disease Models & Mechanisms (2021) 14, dmm047225. doi:10.1242/dmm.047225 RESEARCH ARTICLE Hearing impairment due to Mir183/96/182 mutations suggests both loss-of-function and gain-of-function effects Morag A. Lewis1,2,*, Francesca Di Domenico1, Neil J. Ingham1,2, Haydn M. Prosser2 and Karen P. Steel1,2 ABSTRACT birth. However, this is not the cause of the hearing loss; even before The microRNA miR-96 is important for hearing, as point mutations in the onset of normal hearing, homozygote hair cells fail to mature humans and mice result in dominant progressive hearing loss. Mir96 both morphologically and physiologically, remaining in their is expressed in sensory cells along with Mir182 and Mir183, but the immature state, and heterozygote hair cells show a developmental roles of these closely-linked microRNAs are as yet unknown. Here, delay. miR-96 is thus thought to be responsible for coordinating we analyse mice carrying null alleles of Mir182, and of Mir183 and hair cell maturation (Chen et al., 2014; Kuhn et al., 2011). Mir96 together to investigate their roles in hearing. We found that Overexpression of the three miRNAs also results in cochlear defects Mir183/96 heterozygous mice had normal hearing and homozygotes and hearing loss (Weston et al., 2018). The complete loss of all were completely deaf with abnormal hair cell stereocilia bundles and mature miRNAs from the inner ear results in early developmental reduced numbers of inner hair cell synapses at 4 weeks of age. defects including a severely truncated cochlear duct (Friedman Mir182 knockout mice developed normal hearing then exhibited et al., 2009; Soukup et al., 2009). -

Timo Lassmann, Yoshiko Maida 2, Yasuhiro Tomaru, Mami Yasukawa
Int. J. Mol. Sci. 2015, 16, 1192-1208; doi:10.3390/ijms16011192 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article Telomerase Reverse Transcriptase Regulates microRNAs Timo Lassmann 1,†, Yoshiko Maida 2,†, Yasuhiro Tomaru 1, Mami Yasukawa 2, Yoshinari Ando 1, Miki Kojima 1, Vivi Kasim 2, Christophe Simon 1, Carsten O. Daub 1, Piero Carninci 1, Yoshihide Hayashizaki 1,* and Kenkichi Masutomi 2,3,* 1 RIKEN Omics Science Center, RIKEN Yokohama Institute, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan; E-Mails: [email protected] (T.L.); [email protected] (Y.T.); [email protected] (Y.A.); [email protected] (M.K.); [email protected] (C.S.); [email protected] (C.O.D.); [email protected] (P.C.) 2 Cancer Stem Cell Project, National Cancer Center Research Institute, 5-1-1 Tsukiji, Chuo-ku, Tokyo 104-0045, Japan; E-Mails: [email protected] (Y.M.); [email protected] (M.Y.); [email protected] (V.K.) 3 Precursory Research for Embryonic Science and Technology (PREST), Japan Science and Technology Agency, 4-1-8 Honcho Kawaguchi, Saitama 332-0012, Japan † These authors contributed equally to this work. * Authors to whom correspondence should be addressed; E-Mails: [email protected] (Y.H.); [email protected] (K.M.); Tel.: +81-45-503-9248 (Y.H.); +81-3-3547-5173 (K.M.); Fax: +81-45-503-9216 (Y.H.); +81-3-3547-5123 (K.M.). Academic Editor: Martin Pichler Received: 22 November 2014 / Accepted: 26 December 2014 / Published: 6 January 2015 Abstract: MicroRNAs are small non-coding RNAs that inhibit the translation of target mRNAs. -
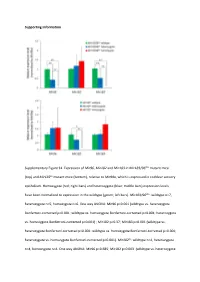
Supporting Information Supplementary Figure S1
Supporting Information Supplementary Figure S1. Expression of Mir96, Mir182 and Mir183 in Mir183/96dko mutant mice (top) and Mir182ko mutant mice (bottom), relative to Mir99a, which is expressed in cochlear sensory epithelium. Homozygote (red; right bars) and heterozygote (blue; middle bars) expression levels have been normalised to expression in the wildtype (green; left bars). Mir183/96dko: wildtype n=7, heterozygote n=5, homozygote n=6. One way ANOVA: Mir96 p<0.001 (wildtype vs. heterozygote Bonferroni‐corrected p<0.001; wildtype vs. homozygote Bonferroni‐corrected p<0.001; heterozygote vs. homozygote Bonferroni‐corrected p=0.001) ; Mir182 p=0.37; Mir183 p<0.001 (wildtype vs. heterozygote Bonferroni‐corrected p=0.001; wildtype vs. homozygote Bonferroni‐corrected p<0.001; heterozygote vs. homozygote Bonferroni‐corrected p<0.001). Mir182ko: wildtype n=4, heterozygote n=4, homozygote n=4. One way ANOVA: Mir96 p=0.685; Mir182 p=0.003 (wildtype vs. heterozygote Bonferroni‐corrected p=0.397; wildtype vs. homozygote Bonferroni‐corrected p=0.003; heterozygote vs. homozygote Bonferroni‐corrected p=0.032); Mir183 p=0.04 (wildtype vs. heterozygote Bonferroni‐corrected p=1.0; wildtype vs. homozygote Bonferroni‐corrected p=0.068; heterozygote vs. homozygote Bonferroni‐corrected p=0.094), Error bars are standard deviation (* = P < 0.05, ** = P ≤ 0.01). Supplementary Figure S2. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir183/96dko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. Supplementary Figure S3. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir182ko mice at all ages tested. -

Language Impairment Resulting from a De Novo Deletion of 7Q32.1-Q33: a Case
bioRxiv preprint doi: https://doi.org/10.1101/047241; this version posted April 6, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Language impairment resulting from a de novo deletion of 7q32.1-q33: a case report Mª Salud Jiménez-Romero, Montserrat Barcos-Martínez, Isabel Espejo-Portero, Antonio Benítez-Burraco Corresponding author: Antonio Benítez-Burraco, PhD. Department of Spanish Philology and its Didactics, University of Huelva, Huelva, Spain e-mail: [email protected] Affiliations for the other authors: Mª Salud Jiménez-Romero: Department of Psychology, University of Córdoba, Córdoba, Spain & Maimónides Institute of Biomedical Research, Córdoba, Spain Montserrat Barcos-Martínez: Laboratory of Molecular Genetics, University Hospital “Reina Sofía”, Córdoba, Spain &Maimónides Institute of Biomedical Research, Córdoba, Spain Isabel Espejo-Portero: Laboratory of Molecular Genetics, University Hospital “Reina Sofía”, Córdoba, Spain & Maimónides Institute of Biomedical Research, Córdoba, Spain bioRxiv preprint doi: https://doi.org/10.1101/047241; this version posted April 6, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 2 ABSTRACT Chromosome 7 is a hot spot for cognitive disorders involving language deficits. We report on a girl who presents with a cognitive and speech delay, motor problems, hearing loss, and behavioral disturbances, and a de novo deletion within 7q32.1-q33 (chromosome position: chr7:127109685-132492196, hg 18). Several genes involved in brain development and function are located within the deleted region. -

Noncoding Rnas As Novel Pancreatic Cancer Targets
NONCODING RNAS AS NOVEL PANCREATIC CANCER TARGETS by Amy Makler A Thesis Submitted to the Faculty of The Charles E. Schmidt College of Science In Partial Fulfillment of the Requirements for the Degree of Master of Science Florida Atlantic University Boca Raton, FL August 2018 Copyright 2018 by Amy Makler ii ACKNOWLEDGEMENTS I would first like to thank Dr. Narayanan for his continuous support, constant encouragement, and his gentle, but sometimes critical, guidance throughout the past two years of my master’s education. His faith in my abilities and his belief in my future success ensured I continue down this path of research. Working in Dr. Narayanan’s lab has truly been an unforgettable experience as well as a critical step in my future endeavors. I would also like to extend my gratitude to my committee members, Dr. Binninger and Dr. Jia, for their support and suggestions regarding my thesis. Their recommendations added a fresh perspective that enriched our initial hypothesis. They have been indispensable as members of my committee, and I thank them for their contributions. My parents have been integral to my successes in life and their support throughout my education has been crucial. They taught me to push through difficulties and encouraged me to pursue my interests. Thank you, mom and dad! I would like to thank my boyfriend, Joshua Disatham, for his assistance in ensuring my writing maintained a logical progression and flow as well as his unwavering support. He was my rock when the stress grew unbearable and his encouraging words kept me pushing along.