2013年12月~2014年11月末 報告入手分)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Material Computational Prediction of SARS
Supplementary_Material Computational prediction of SARS-CoV-2 encoded miRNAs and their putative host targets Sheet_1 List of potential stem-loop structures in SARS-CoV-2 genome as predicted by VMir. Rank Name Start Apex Size Score Window Count (Absolute) Direct Orientation 1 MD13 2801 2864 125 243.8 61 2 MD62 11234 11286 101 211.4 49 4 MD136 27666 27721 104 205.6 119 5 MD108 21131 21184 110 204.7 210 9 MD132 26743 26801 119 188.9 252 19 MD56 9797 9858 128 179.1 59 26 MD139 28196 28233 72 170.4 133 28 MD16 2934 2974 76 169.9 71 43 MD103 20002 20042 80 159.3 403 46 MD6 1489 1531 86 156.7 171 51 MD17 2981 3047 131 152.8 38 87 MD4 651 692 75 140.3 46 95 MD7 1810 1872 121 137.4 58 116 MD140 28217 28252 72 133.8 62 122 MD55 9712 9758 96 132.5 49 135 MD70 13171 13219 93 130.2 131 164 MD95 18782 18820 79 124.7 184 173 MD121 24086 24135 99 123.1 45 176 MD96 19046 19086 75 123.1 179 196 MD19 3197 3236 76 120.4 49 200 MD86 17048 17083 73 119.8 428 223 MD75 14534 14600 137 117 51 228 MD50 8824 8870 94 115.8 79 234 MD129 25598 25642 89 115.6 354 Reverse Orientation 6 MR61 19088 19132 88 197.8 271 10 MR72 23563 23636 148 188.8 286 11 MR11 3775 3844 136 185.1 116 12 MR94 29532 29582 94 184.6 271 15 MR43 14973 15028 109 183.9 226 27 MR14 4160 4206 89 170 241 34 MR35 11734 11792 111 164.2 37 52 MR5 1603 1652 89 152.7 118 53 MR57 18089 18132 101 152.7 139 94 MR8 2804 2864 122 137.4 38 107 MR58 18474 18508 72 134.9 237 117 MR16 4506 4540 72 133.8 311 120 MR34 10010 10048 82 132.7 245 133 MR7 2534 2578 90 130.4 75 146 MR79 24766 24808 75 127.9 59 150 MR65 21528 21576 99 127.4 83 180 MR60 19016 19049 70 122.5 72 187 MR51 16450 16482 75 121 363 190 MR80 25687 25734 96 120.6 75 198 MR64 21507 21544 70 120.3 35 206 MR41 14500 14542 84 119.2 94 218 MR84 26840 26894 108 117.6 94 Sheet_2 List of stable stem-loop structures based on MFE. -

(12) United States Patent (10) Patent No.: US 9,476,099 B2 Spinella Et Al
US009476.099B2 (12) United States Patent (10) Patent No.: US 9,476,099 B2 Spinella et al. (45) Date of Patent: Oct. 25, 2016 (54) METHOD FOR DETERMINING FOREIGN PATENT DOCUMENTS SENSTIVITY TO DECTABINE WO WO 2012/031008 A2 3, 2012 TREATMENT WO WO 2012, 11885.6 A1 9, 2012 (71) Applicant: TRUSTEES OF DARTMOUTH COLLEGE, Hanover, NH (US) OTHER PUBLICATIONS (72) Inventors: Michael Spinella, Hanover, NH (US); Tsai et al Cancer Cell. Mar. 20, 2012. 21(3):43.0-446 and Supple Maroun J. Beyrouthy, Lebanon, NH mental pages 1-18. (US) Agilent Technologies. DNA Oligo Microarray Gene Lists and Annotations, available via url: <chem.agilent.com/cag?bSp? gene (73) Assignee: Trustees of Dartmouth College, lists.asp printed on Mar. 1, 2016.* Hanover, NH (US) Abele et al. “The EORTC Early Clinical Trials Cooperative Group Experience with 5-Aza-2'-deoxycytidine (NSC 127716) in Patients (*) Notice: Subject to any disclaimer, the term of this with Colo-rectal, Head and Neck, Renal Carcinomas and Malignant patent is extended or adjusted under 35 Melanomas' European Journal of Cancer and Clinical Oncology U.S.C. 154(b) by 0 days. 1987 23(12): 1921-1924. Adewumi et al. “Characterization of Human Embryonic Stem Cell (21) Appl. No.: 14/416,142 Lines by the International StemCell Initiative” Nature Biotechnol ogy 2007 vol. 25(7):803-816. (22) PCT Filed: Jul. 31, 2013 Al-Hajj et al. "Prospective Identification of Tumorigenic Breast Cancer Cells' Proceedings of the National Academy of Sciences (86). PCT No.: PCT/US2013/052899 2003 100(7):3983-3988 with correction. -

Strand Breaks for P53 Exon 6 and 8 Among Different Time Course of Folate Depletion Or Repletion in the Rectosigmoid Mucosa
SUPPLEMENTAL FIGURE COLON p53 EXONIC STRAND BREAKS DURING FOLATE DEPLETION-REPLETION INTERVENTION Supplemental Figure Legend Strand breaks for p53 exon 6 and 8 among different time course of folate depletion or repletion in the rectosigmoid mucosa. The input of DNA was controlled by GAPDH. The data is shown as ΔCt after normalized to GAPDH. The higher ΔCt the more strand breaks. The P value is shown in the figure. SUPPLEMENT S1 Genes that were significantly UPREGULATED after folate intervention (by unadjusted paired t-test), list is sorted by P value Gene Symbol Nucleotide P VALUE Description OLFM4 NM_006418 0.0000 Homo sapiens differentially expressed in hematopoietic lineages (GW112) mRNA. FMR1NB NM_152578 0.0000 Homo sapiens hypothetical protein FLJ25736 (FLJ25736) mRNA. IFI6 NM_002038 0.0001 Homo sapiens interferon alpha-inducible protein (clone IFI-6-16) (G1P3) transcript variant 1 mRNA. Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 GALNTL5 NM_145292 0.0001 (GALNT15) mRNA. STIM2 NM_020860 0.0001 Homo sapiens stromal interaction molecule 2 (STIM2) mRNA. ZNF645 NM_152577 0.0002 Homo sapiens hypothetical protein FLJ25735 (FLJ25735) mRNA. ATP12A NM_001676 0.0002 Homo sapiens ATPase H+/K+ transporting nongastric alpha polypeptide (ATP12A) mRNA. U1SNRNPBP NM_007020 0.0003 Homo sapiens U1-snRNP binding protein homolog (U1SNRNPBP) transcript variant 1 mRNA. RNF125 NM_017831 0.0004 Homo sapiens ring finger protein 125 (RNF125) mRNA. FMNL1 NM_005892 0.0004 Homo sapiens formin-like (FMNL) mRNA. ISG15 NM_005101 0.0005 Homo sapiens interferon alpha-inducible protein (clone IFI-15K) (G1P2) mRNA. SLC6A14 NM_007231 0.0005 Homo sapiens solute carrier family 6 (neurotransmitter transporter) member 14 (SLC6A14) mRNA. -

A Bacterial Protein Targets the BAHD1 Chromatin Complex to Stimulate Type III Interferon Response
A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response Alice Lebreton, Goran Lakisic, Viviana Job, Lauriane Fritsch, To Nam Tham, Ana Camejo, Pierre-Jean Matteï, Béatrice Regnault, Marie-Anne Nahori, Didier Cabanes, et al. To cite this version: Alice Lebreton, Goran Lakisic, Viviana Job, Lauriane Fritsch, To Nam Tham, et al.. A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science, American Association for the Advancement of Science, 2011, 331 (6022), pp.1319-21. 10.1126/sci- ence.1200120. cea-00819299 HAL Id: cea-00819299 https://hal-cea.archives-ouvertes.fr/cea-00819299 Submitted on 26 Jul 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Lebreton et al. Science 2011 doi:10.1126/science.1200120 A Bacterial Protein Targets the BAHD1 Chromatin Complex to Stimulate Type III Interferon Response Alice Lebreton1,2,3, Goran Lakisic4, Viviana Job5, Lauriane Fritsch6, To Nam Tham1,2,3, Ana Camejo7, Pierre-Jean Matteï5, Béatrice Regnault8, Marie-Anne Nahori1,2,3, Didier Cabanes7, Alexis Gautreau4, Slimane Ait-Si-Ali6, Andréa Dessen5, Pascale Cossart1,2,3* and Hélène Bierne1,2,3* 1. -

Genomic Architecture of Histone 3 Lysine 27 Trimethylation During Late Ovine Skeletal Muscle Development
doi: 10.1111/age.12145 Genomic architecture of histone 3 lysine 27 trimethylation during late ovine skeletal muscle development † ‡ K. Byrne*1, S. McWilliam*1, T. Vuocolo*, C. Gondro , N. E. Cockett and R. L. Tellam* † *CSIRO Animal, Food and Health Sciences, Queensland Bioscience Precinct, 306 Carmody Rd, St Lucia, QLD, 4067, Australia. School of ‡ Environmental and Rural Science, University of New England, Armidale, NSW, Australia. Department of Animal, Dairy and Veterinary Sciences, Utah State University, Logan, UT, USA. Summary The ruminant developmental transition from late foetus to lamb is associated with marked changes in skeletal muscle structure and function that reflect programming for new physiological demands following birth. To determine whether epigenetic changes are involved in this transition, we investigated the genomic architecture of the chromatin modification, histone 3 lysine 27 trimethylation (H3K27me3), which typically regulates early life developmental processes; however, its role in later life processes is unclear. Chromatin immunoprecipitation coupled with next-generation sequencing was used to map H3K27me3 nucleosomes in ovine longissimus lumborum skeletal muscle at 100 days of gestation and 12 weeks post-partum. In both states, H3K27me3 modification was associated with genes, transcription start sites and CpG islands and with transcriptional silencing. The H3K27me3 peaks consisted of two major categories, promoter specific and regional, with the latter the dominant feature. Genes encoding homeobox transcription factors regulating early life development and genes involved in neural functions, particularly gated ion channels, were strongly modified by H3K27me3. Gene promoters differentially modified by H3K27me3 in the foetus and lamb were enriched for gated ion channels, which may reflect changes in neuromuscular function. -

Directed Differentiation of Human Embryonic Stem Cells Into Haematopoietic and Definitive Endodermal Lineages
DIRECTED DIFFERENTIATION OF HUMAN EMBRYONIC STEM CELLS INTO HAEMATOPOIETIC AND DEFINITIVE ENDODERMAL LINEAGES ABRAHAM SUMAN MARY (M.Sc MICROBIOLOGY, UNIV. OF MUMBAI, INDIA) A THESIS SUBMITTED FOR THE DEGREE OF MASTER OF SCIENCE DEPARTMENT OF BIOCHEMISTRY NATIONAL UNIVERSITY OF SINGAPORE 2009 ACKNOWLEDGEMENTS In all things I give YOU glory! You have always led me through amazing paths and given me gifts that I don’t deserve. I thank you Lord for all the blessings you constantly shower on me. Everything is possible with God! I thank Dr. Alan Colman for being my guide and helping me to initiate the work contained in this dissertation. His support and encouragement have been invaluable. Thank you, Alan for your support through the years. Dr. Norris Ray Dunn took me under his wing and guided me through this endeavour. He has been a true mentor, always willing to teach, and I have learned a lot from him. Ray, thank you for showing me the way and helping me reach this juncture. A significant part of the research was conducted at ES Cell International Pte Ltd to whom I would like to express my sincere gratitude. Triona, Jacqui, Robert, Michael, Bruce, Chirag, Suzan and so many others have played important roles and encouraged me at all times. Critical portions of this work were done at the Institute of Medical Biology, A*Star. I would like to register my appreciation for the support and help provided by many people in IMB especially, members of the Ray Dunn lab, Mike Jones lab and Alan Colman lab. Kee Yew, thank you for giving me your precious time and helping me with some of the most important data in this dissertation. -

SF3B1-Mutated Chronic Lymphocytic Leukemia Shows Evidence Of
SF3B1-mutated chronic lymphocytic leukemia shows evidence of NOTCH1 pathway activation including CD20 downregulation by Federico Pozzo, Tamara Bittolo, Erika Tissino, Filippo Vit, Elena Vendramini, Luca Laurenti, Giovanni D'Arena, Jacopo Olivieri, Gabriele Pozzato, Francesco Zaja, Annalisa Chiarenza, Francesco Di Raimondo, Antonella Zucchetto, Riccardo Bomben, Francesca Maria Rossi, Giovanni Del Poeta, Michele Dal Bo, and Valter Gattei Haematologica 2020 [Epub ahead of print] Citation: Federico Pozzo, Tamara Bittolo, Erika Tissino, Filippo Vit, Elena Vendramini, Luca Laurenti, Giovanni D'Arena, Jacopo Olivieri, Gabriele Pozzato, Francesco Zaja, Annalisa Chiarenza, Francesco Di Raimondo, Antonella Zucchetto, Riccardo Bomben, Francesca Maria Rossi, Giovanni Del Poeta, Michele Dal Bo, and Valter Gattei SF3B1-mutated chronic lymphocytic leukemia shows evidence of NOTCH1 pathway activation including CD20 downregulation. Haematologica. 2020; 105:xxx doi:10.3324/haematol.2020.261891 Publisher's Disclaimer. E-publishing ahead of print is increasingly important for the rapid dissemination of science. Haematologica is, therefore, E-publishing PDF files of an early version of manuscripts that have completed a regular peer review and have been accepted for publication. E-publishing of this PDF file has been approved by the authors. After having E-published Ahead of Print, manuscripts will then undergo technical and English editing, typesetting, proof correction and be presented for the authors' final approval; the final version of the manuscript will -
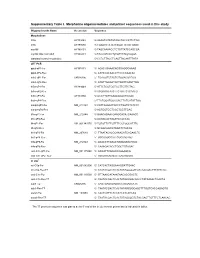
Supplementary Table I. Morpholino Oligonucleotides and Primer Sequences Used in This Study
Supplementary Table I. Morpholino oligonucleotides and primer sequences used in this study Oligonucleotide Name Accession Sequence Morpholinos tlr5a AY389449 5'-AAAGTGTATGTAGCTGCCATTCTGG tlr5b AY389450 5'-TGAATGTATATCCCATTCTGTGAGC myd88 AY388401 5'-TAGCAAAACCTCTGTTATCCAGCGA myd88 5bp mismatch AY388401 5'-TAcCAtAACCTgTGTTATCgAGgGA standard control morpholino 5'-CCTCTTACCTCAGTTACAATTTATA qRT-PCR ppial-qP1-Fw AY391451 5’- ACACTGAAACACGGAGGCAAAG ppial-qP2-Rev 5’- CATCCACAACCTTCCCGAACAC irak3-qP1-Fw CK026195 5’- TGAGGTCTACTGTGGACGATGG irak3-qP2-Rev 5’- ATGTTAGGATGCTGGTTGAGTTGG tlr5a-qP1-Fw AY389449 5’-ATTCTGGTGGTGCTTGTTGTAG tlr5a-qP2-Rev 5’-ACGAGGTAACTTCTGTTCTCAATG tlr5b-qP3-Fw AY389450 5’-GCGTTGTTGAAGAGGCTGGAC tlr5b-qP4-Rev 5’-TTCTGGATGGCCACTTCTCATATTGG mmp9-qP3-Fw NM_213123 5’-CATTAAAGATGCCCTGATGTATCCC mmp9-qP4-Rev 5’-AGTGGTGGTCCGTGGTTGAG il1b-qP1-Fw NM_212844 5’-GAACAGAATGAAGCACATCAAACC il1b-qP2-Rev 5’-ACGGCACTGAATCCACCAC il8-qP1-Fw XM_001342570 5’-TGTGTTATTGTTTTCCTGGCATTTC il8-qP2-Rev 5’-GCGACAGCGTGGATCTACAG ifn1-qP3-Fw NM_207640 5’- TTAATACACGCAAAGATGAGAACTC ifn1-qP4-Rev 5’- GCCAAGCCATTCGCAAGTAG tnfa-qP5-Fw NM_212829 5’- AGACCTTAGACTGGAGAGATGAC tnfa-qP6-Rev 5’- CAAAGACACCTGGCTGTAGAC cxcl-C1c-qP1-Fw NM_001115060 5’- GGCATTCACACCCAAAGCG cxcl-C1c-qP2_Rev 5’- GCGAGCACGATTCACGAGAG * In situ ccl-C5a-Fw NM_001082906 5’- CATCACTAGGAAAGGATTGAAC ccl-C5a-Rev-T7 5’- TAATACGACTCACTATAGGGGATGTCAAAGACTTTATTCAC cxcl-C1c-Fw NM_001115060 5’- GTTAAACATAAATAACACCGACTC cxcl-C1c-Rev-T7 5’- TAATACGACTCACTATAGGGACACCCTATAAAACTGAGTA irak3-Fw CK026195 5’- CAGTGAGAGAGGCATGAAACATC -

Supplementary Material Study Sample the Vervets Used in This
Supplementary Material Study sample The vervets used in this study are part of a pedigreed research colony that has included more than 2,000 monkeys since its founding. Briefly, the Vervet Research Colony (VRC) was established at UCLA during the 1970’s and 1980’s from 57 founder animals captured from wild populations on the adjacent Caribbean islands of St. Kitts and Nevis; Europeans brought the founders of these populations to the Caribbean from West Africa in the 17th Century 1. The breeding strategy of the VRC has emphasized the promotion of diversity, the preservation of the founding matrilines, and providing all females and most of the males an opportunity to breed. The colony design modeled natural vervet social groups to facilitate behavioral investigations in semi-natural conditions. Social groups were housed in large outdoor enclosures with adjacent indoor shelters. Each enclosure had chain link siding that provided visual access to the outside, with one or two large sitting platforms and numerous shelves, climbing structures and enrichments devices. The monkeys studied were members of 16 different social matrilineal groups, containing from 15 to 46 members per group. In 2008 the VRC was moved to Wake Forest School of Medicine’s Center for Comparative Medicine Research, however the samples for gene expression measurements in Dataset 1 (see below) and the MRI assessments used in this study occurred when the colony was at UCLA. Gene expression phenotypes Two sets of gene expression measurements were collected. Dataset 1 used RNA obtained from whole blood in 347 vervets, assayed by microarray (Illumina HumanRef-8 v2); Dataset 2 assayed gene expression by RNA-Seq, in RNA obtained from 58 animals, with seven tissues (adrenal, blood, Brodmann area 46 [BA46], caudate, fibroblast, hippocampus and pituitary) measured in each animal. -

Highly Conserved Upstream Sequences for Transcription Factor Genes and Implications for the Regulatory Network
Highly conserved upstream sequences for transcription factor genes and implications for the regulatory network Hisakazu Iwama*† and Takashi Gojobori*‡§ *Center for Information Biology and DNA Data Bank of Japan, National Institute of Genetics, Research Organization of Information and Systems, Yata 1111, Mishima, 411-8540 Japan; and ‡Integrated Database Group, Biological Information Research Center, National Institute of Advanced Industrial Science and Technology, Time 24 Building, 10th Floor, 2-45 Aomi, Koto-ku, Tokyo 135-0064, Japan Communicated by Wen-Hsiung Li, University of Chicago, Chicago, IL, October 15, 2004 (received for review May 27, 2004) Identifying evolutionarily conserved blocks in orthologous We report here that the genes with high upstream conserva- genomic sequences is an effective way to detect regulatory ele- tion are predominantly transcription factor (TF) genes. Further- ments. In this study, with the aim of elucidating the architecture of more, we show that the developmental process-related TF genes the regulatory network, we systematically estimated the degree of have significantly higher conservation of the upstream sequences conservation of the upstream sequences of 3,750 human–mouse than other TF genes. orthologue pairs along 8-kb stretches. We found that the genes with high upstream conservation are predominantly transcription Materials and Methods factor (TF) genes. In particular, developmental process-related TF Orthologue Identification and Upstream Sequence Collection. We genes showed significantly higher -

Identifying Changes in Punitive Transcriptional Factor Binding Sites Created by Pparα/Δ/Γ Snps Associated with Disease
Journal of Biosciences and Medicines, 2017, 5, 81-100 http://www.scirp.org/journal/jbm ISSN Online: 2327-509X ISSN Print: 2327-5081 Identifying Changes in Punitive Transcriptional Factor Binding Sites Created by PPARα/δ/γ SNPs Associated with Disease Norman E. Buroker Department of Pediatrics, University of Washington, Seattle, WA, USA How to cite this paper: Buroker, N.E. Abstract (2017) Identifying Changes in Punitive Transcriptional Factor Binding Sites Single nucleotide polymorphisms (SNPs) located in the PPARα/δ/γ genes that Created by PPARα/δ/γ SNPs Associated have been previously found to be significantly associated with human disease with Disease. Journal of Biosciences and or a condition were also found to alter the genes punitive transcriptional fac- Medicines, 5, 81-100. tor binding sites (TFBS). Two alleles (C/G) of the PPARα SNP (rs1800206) https://doi.org/10.4236/jbm.2017.54008 were found to generate 7 common and 8 unique punitive TFBS. One of the Received: March 29, 2017 unique TFBS created by the minor G (0.02) allele is for the T-Box 4 (TBX4) Accepted: April 27, 2017 transcription factor which is associated with heritable pulmonary arterial Published: April 30, 2017 hypertension. Two alleles (A/G) of the PPARδ SNP (rs2016520) were found to Copyright © 2017 by author and generate 20 unique punitive TFBS while the two alleles (C/G) of the PPARδ Scientific Research Publishing Inc. SNP (rs9794) were found to generate 11 common and 11unique punitive This work is licensed under the Creative TFBS. The alleles of the PPARγ SNPs (rs10865710, rs12629751, rs709158, Commons Attribution International rs1805192 and rs3856806) were found to generate 15, 12, 16, 2 and 21 com- License (CC BY 4.0). -

Whole-Transcriptome Analysis of Fully Viable Energy Efficient Glycolytic
CANCER GENOMICS & PROTEOMICS 17 : 469-497 (2020) doi:10.21873/cgp.20205 Whole-transcriptome Analysis of Fully Viable Energy Efficient Glycolytic-null Cancer Cells Established by Double Genetic Knockout of Lactate Dehydrogenase A/B or Glucose-6-Phosphate Isomerase ELIZABETH MAZZIO 1, RAMESH BADISA 1, NZINGA MACK 1, SHAMIR CASSIM 2, MASA ZDRALEVIC 3# , JACQUES POUYSSEGUR 2,3 and KARAM F.A. SOLIMAN 1 1College of Pharmacy & Pharmaceutical Sciences, Florida A&M University, Tallahassee, FL, U.S.A.; 2Department of Medical Biology, Centre Scientifique de Monaco, Monaco, Monaco; 3University Côte d'Azur, IRCAN, CNRS, Centre A. Lacassagne, Nice, France Abstract. Background/Aim: Nearly all mammalian tumors viability. These findings show a high biochemical energy of diverse tissues are believed to be dependent on efficiency as measured by ATP in glycolysis-null cells. Next, fermentative glycolysis, marked by elevated production of transcriptomic analysis conducted on 48,226 mRNA lactic acid and expression of glycolytic enzymes, most notably transcripts reflect 273 differentially expressed genes (DEGS) lactic acid dehydrogenase (LDH). Therefore, there has been in the GPI KO clone set, 193 DEGS in the LDHA/B DKO significant interest in developing chemotherapy drugs that clone set with 47 DEGs common to both KO clones. selectively target various isoforms of the LDH enzyme. Glycolytic-null cells reflect up-regulation in gene transcripts However, considerable questions remain as to the typically associated with nutrient deprivation / fasting and consequences of biological ablation of LDH or upstream possible use of fats for energy: thioredoxin interacting protein targeting of the glycolytic pathway. Materials and Methods: (TXNIP), mitochondrial 3-hydroxy-3-methylglutaryl-CoA In this study, we explore the biochemical and whole synthase 2 (HMGCS2), PPAR γ coactivator 1 α ( PGC-1 α), transcriptomic effects of CRISPR-Cas9 gene knockout (KO) and acetyl-CoA acyltransferase 2 (ACAA2).