Additional File 1 List of Transcripts
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

NVND-2019 Book
2nd International Conference on Neurovascular and Neurodegenerative Diseases October 28-30 2019 Media Partner Venue Paris Marriott Charles de Gaulle Airport Hotel 5 Allee du Verger, Zone Hoteliere Roissy en France, 95700 France Day- 1 Monday | October 28, 2019 Keynote Presentations Cerebrovascular Lesions in Pick Complex Diseases: A Neuropathological Study with a 7.0-tesla Magnetic Resonance Imaging Jacques De Reuck Université Lille 2, INSERM U1171, Degenerative & Vascular Cognitive Disorders, CHR Lille, France Abstract Introduction: Pick complex refers to a spectrum of diseases that have in common the presence of tau inclusions. The main neuropathological phenotypes comprise tau-frontotemporal lobar degeneration (Tau-FTLD), progressive supranuclear palsy (PSP) and corticobasal degeneration (CBD). The present neuropathological study investigates their incidence of cerebrovascular lesions. Material and Methods: seventy patients underwent an autopsy and post-mortem MRI. The brains consisted of 14 with Tau-FTLD, 22 with PSP, 6 with CBD and 28 controls, who had no history of a brain disease. A whole coronal section of a cerebral hemisphere, at the level of the mamillary body, was taken for the semi-quantitative evaluation of the small cerebrovascular lesions such as white matter changes (WMCs), cortical micro-bleeds (CoMBs), and cortical micro-infarcts (CoMIs). In addition the severity and the distribution of WMCs, CoMIs and CoMBs were examined with 7.0-tesla MRI on three coronal sections of a cerebral hemisphere. Results: on neuropathological examination severe WMCs and more CoMBs are observed in Tau-FTLD, while the latter are also more frequent in CBD. The MRI examination shows that severe WMCs are present in the frontal sections not only of the Tau-FTLD but also to a lesser degree of the PSP and CBD brains. -

MOUSE INTERLEUKIN-28B/INTERFERON-LAMBDA 3, CARRIER FREE Product Number: 12821-1 Lot Number: 5559 Size: 25 Μg
MOUSE INTERLEUKIN-28B/INTERFERON-LAMBDA 3, CARRIER FREE Product Number: 12821-1 Lot Number: 5559 Size: 25 µg Description: Mouse Interleukin-28B/Interferon-lambda 3, Carrier Free Source: A DNA sequence encoding the mature mouse IL-28B/IFN-λ3 (Asp 20 - Val 193) (Kotenko, S.V. et al., 2003, Nat. Immunol. 4(1):69 - 77) was expressed in E. coli. Form: Lyophilized Buffer: Phosphate-buffered saline (PBS) Reconstitution: It is recommended that sterile PBS be added to the vial to prepare a stock solution of no less than 100 μg/mL. Endotoxin: < 1 EU/µg Molecular Weight: The 175 amino acid residue methionyl form of recombinant mouse IL-28B has a predicted molecular mass of approximately 19.7 kDa. Purity: > 95% Synonyms: Mu IL-28B; Mu IFN-λ3 Accession #: NP_796370 Assays Used to Measure Bioactivity: Human HepG2 cells infected with encephalomyocarditis virus (Sheppard, P. et al., 2003, Nature Immunol. 4:63). The ED50 for this effect is typically 7.5 - 37.5 ng/mL. Shipping Conditions: Wet Ice Physical State of Product During Shipping: Lyophilized Special Conditions/Comments: After receipt, this product should be kept at -70˚C or below for retention of full activity. Upon reconstitution, this cytokine can be stored under sterile conditions at 2˚C to 8˚C for one month or at -20˚C to -70˚C in a manual defrost freezer for three months without detection loss of activity. Avoid repeated freeze-thaw cycles. For more information on protein handling, visit the PBL website at www.interferonsource.com . Product Information: Human IL-28A, IL-28B, and IL-29, also named interferon-λ2 (IFN-λ2), IFN-λ3, and IFN-λ1, respectively, are newly identified class II cytokine receptor ligands that are distantly related to members of the IL-10 family (11- 13% aa sequence identity) and type I IFN family (15 - 19% aa sequence identity).1 – 3 The genes encoding these three cytokines are localized to chromosome 19 and each is composed of multiple exons. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Kinesin Family Member 6 (Kif6) Is Necessary for Spine Development in Zebrafish 8 9 10 11 12 Jillian G
Page 1 of 40 Developmental Dynamics 1 2 3 4 5 6 7 Kinesin family member 6 (kif6) is necessary for spine development in zebrafish 8 9 10 11 12 Jillian G. Buchan1, Ryan S. Gray2, John M. Gansner6, David M. Alvarado3, Lydia Burgert4, 13 14 Jonathan D. Gitlin7, Christina A. Gurnett3,4,5, Matthew I. Goldsmith1,4,* 15 16 17 Departments of 1Genetics, 2Developmental Biology, 3Orthopaedic Surgery, 4Pediatrics, 18 5 6 19 Neurology, Washington University School of Medicine, St. Louis, MO, 63110; Dana-Farber 20 Cancer Institute, Boston, MA, 02215; 7Eugene Bell Center for Regenerative Biology, Marine 21 Biological Laboratory, Woods Hole, MA, 02543 22 23 24 25 26 *Corresponding author: Matthew I. Goldsmith, MD 27 28 Department of Pediatrics 29 Washington University School of Medicine 30 [email protected] 31 660 S Euclid Ave, Campus Box 8208 32 St Louis, MO 63110 33 Phone: (314) 286-2769 34 35 Fax: (314) 286-2784 36 37 38 39 Running Title: Kif6 in zebrafish spine development 40 41 Keywords: kinesin, scoliosis, Danio rerio 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 Developmental Dynamics Page 2 of 40 1 2 3 ABSTRACT 4 5 6 Background: Idiopathic scoliosis is a form of spinal deformity that affects 2-3% of children and 7 8 results in curvature of the spine without structural defects of the vertebral units. The 9 10 11 pathogenesis of idiopathic scoliosis remains poorly understood, in part due to the lack of a 12 13 relevant animal model. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Molecular Characterization of Tea Catechin Treated Human Prostate Cancer Cell Lines Yewseok Suh
Florida State University Libraries Electronic Theses, Treatises and Dissertations The Graduate School 2006 Molecular Characterization of Tea Catechin Treated Human Prostate Cancer Cell Lines Yewseok Suh Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] THE FLORIDA STATE UNIVERSITY COLLEGE OF ARTS AND SCIENCES MOLECULAR CHARACTERIZATION OF TEA CATECHIN TREATED HUMAN PROSTATE CANCER CELL LINES By YEWSEOK SUH A Dissertation submitted to the Department of Chemistry and Biochemistry in partial fulfillment of the requirements for the degree of Doctor of Philosophy Degree Awarded: Summer Semester, 2006 The members of the Committee approve the dissertation of Yewseok Suh defended on May 12, 2006. Qing-Xiang Amy Sang Professor Directing Dissertation Thomas C.S. Keller III Outside Committee Member Joseph B. Schlenoff Committee Member Hong Li Committee Member Approved: Naresh Dalal, Chair, Department of Chemistry and Biochemistry Joseph Travis, Dean, College of Arts and Sciences The Office of Graduate Studies has verified and approved the above named committee members. ii This dissertation is dedicated to my parents for their endless love and encouragement, to my lovely wife Inok Park for her support, and to our charming daughter, Tae-won. iii ACKNOWLEDGEMENTS I would like to express my gratitude to my major professor for her support throughout the research. Also thanks to all my committee members, Dr. Thomas C. S. Keller III, Dr. Hong Li, and Dr. Joseph B. Schlenoff, for their support, advice, and guidance. Special thanks to our lab members especially, Ziad Sahab and Robert G. Newcomer and former lab member Douglas R. -

Mutations in Kinesin Family Member 6 Reveal Specific Role in Ependymal Cell Ciliogenesis and Human Neurological Development
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2018 Mutations in Kinesin family member 6 reveal specific oler in ependymal cell ciliogenesis and human neurological development Mia J. Konjikusic Patra Yeetong Curtis W. Boswell Chanjae Lee Elle C. Roberson See next page for additional authors Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Authors Mia J. Konjikusic, Patra Yeetong, Curtis W. Boswell, Chanjae Lee, Elle C. Roberson, Rungnapa Ittiwut, Kanya Suphapeetiporn, Brian Ciruna, Christina A. Gurnett, John B. Wallingford, Vorasuk Shotelersuk, and Ryan S. Gray RESEARCH ARTICLE Mutations in Kinesin family member 6 reveal specific role in ependymal cell ciliogenesis and human neurological development 1,2 3,4,5 6 2 Mia J. KonjikusicID , Patra Yeetong , Curtis W. BoswellID , Chanjae Lee , Elle 2 3,4 3,4 6 C. RobersonID , Rungnapa IttiwutID , Kanya Suphapeetiporn , Brian Ciruna , Christina 7 2 3,4 1 A. Gurnett , John B. Wallingford , Vorasuk Shotelersuk *, Ryan S. GrayID * 1 Department of Pediatrics, Dell Pediatric Research Institute, The University of Texas at Austin, Dell Medical School, Austin, Texas, United States of America, 2 Department of Molecular Biosciences, Patterson Labs, a1111111111 The University of Texas at Austin, Austin, Texas, United States of America, 3 Center of Excellence for a1111111111 Medical Genetics, Department of Pediatrics, Faculty of Medicine, Chulalongkorn University, Bangkok, a1111111111 Thailand, 4 Excellence Center for Medical Genetics, -

Discovery of Candidate DNA Methylation Cancer Driver Genes
Published OnlineFirst May 10, 2021; DOI: 10.1158/2159-8290.CD-20-1334 RESEARCH ARTICLE Discovery of Candidate DNA Methylation Cancer Driver Genes Heng Pan1,2,3, Loïc Renaud4,5,6,7, Ronan Chaligne4,5,6, Johannes Bloehdorn8, Eugen Tausch8, Daniel Mertens9, Anna Maria Fink10, Kirsten Fischer10, Chao Zhang3,6, Doron Betel3,6, Andreas Gnirke11, Marcin Imielinski1,3,4,5,12, Jérôme Moreaux13,14,15,16, Michael Hallek10, Alexander Meissner11,17, Stephan Stilgenbauer8, Catherine J. Wu11,18, Olivier Elemento1,2,3,5, and Dan A. Landau3,4,5,6 Downloaded from cancerdiscovery.aacrjournals.org on September 28, 2021. © 2021 American Association for Cancer Research. Published OnlineFirst May 10, 2021; DOI: 10.1158/2159-8290.CD-20-1334 ABSTRACT Epigenetic alterations, such as promoter hypermethylation, may drive cancer through tumor suppressor gene inactivation. However, we have limited ability to differentiate driver DNA methylation (DNAme) changes from passenger events. We developed DNAme driver inference–MethSig–accounting for the varying stochastic hypermethylation rate across the genome and between samples. We applied MethSig to bisulfite sequencing data of chronic lymphocytic leukemia (CLL), multiple myeloma, ductal carcinoma in situ, glioblastoma, and to methylation array data across 18 tumor types in TCGA. MethSig resulted in well-calibrated quantile–quantile plots and reproducible inference of likely DNAme drivers with increased sensitivity/specificity compared with benchmarked methods. CRISPR/Cas9 knockout of selected candidate CLL DNAme drivers provided a fitness advantage with and without therapeutic intervention. Notably, DNAme driver risk score was closely associated with adverse outcome in independent CLL cohorts. Collectively, MethSig represents a novel inference framework for DNAme driver discovery to chart the role of aberrant DNAme in cancer. -

2018 Chen Lingyan 1448129
This electronic thesis or dissertation has been downloaded from the King’s Research Portal at https://kclpure.kcl.ac.uk/portal/ Genetics and Epigenetics in Systemic Lupus Erythematosus Chen, Lingyan Awarding institution: King's College London The copyright of this thesis rests with the author and no quotation from it or information derived from it may be published without proper acknowledgement. END USER LICENCE AGREEMENT Unless another licence is stated on the immediately following page this work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International licence. https://creativecommons.org/licenses/by-nc-nd/4.0/ You are free to copy, distribute and transmit the work Under the following conditions: Attribution: You must attribute the work in the manner specified by the author (but not in any way that suggests that they endorse you or your use of the work). Non Commercial: You may not use this work for commercial purposes. No Derivative Works - You may not alter, transform, or build upon this work. Any of these conditions can be waived if you receive permission from the author. Your fair dealings and other rights are in no way affected by the above. Take down policy If you believe that this document breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 03. Oct. 2021 GENETICS AND EPIGENETICS IN SYSTEMIC LUPUS ERYTHEMATOSUS Lingyan CHEN Department of Medical & Molecular Genetics Faculty of Life Sciences & Medicine King’s College London This thesis is submitted for the degree of DoCtor of Philosophy (PhD) 1 To my parents 2 DeClaration The work described in this thesis was carried out within the Vyse Immunogenetics Group in the Department of Medical and Molecular Genetics at King’s College London under the supervision of Professor Timothy James Vyse and Dr David Lester Morris and between the years of October 2014 and March 2018. -
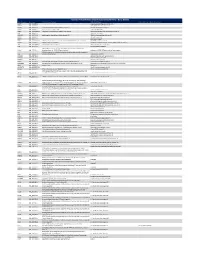
Ncounter® Autoimmune Discovery Consortium Panel
nCounter® Autoimmune Discovery Consortium Panel - Gene Details Official Symbol Accession Alias / Previous Symbol Official Full Name Other targets or Isoform Information AAMP NM_001087.3 angio associated migratory cell protein ABHD6 NM_020676.5 abhydrolase domain containing 6 ACKR2 NM_001296.3 CMKBR9,CCBP2;chemokine binding protein 2 atypical chemokine receptor 2 ACOXL NM_018308.1 acyl-Coenzyme A oxidase-like acyl-CoA oxidase like ACSL6 NM_001009185.1 FACL6;fatty-acid-Coenzyme A ligase, long-chain 6 acyl-CoA synthetase long chain family member 6 ADA NM_000022.2 adenosine deaminase ADAM30 NM_021794.2 a disintegrin and metalloproteinase domain 30 ADAM metallopeptidase domain 30 ADCY3 NM_004036.3 adenylate cyclase 3 ADCY7 NM_001114.4 adenylate cyclase 7 AFF3 NM_001025108.1 LAF4;lymphoid nuclear protein related to AF4,AF4/FMR2 family, member 3 AF4/FMR2 family member 3 AGAP2 NM_014770.3 CENTG1;centaurin, gamma 1 ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 AHI1 NM_001134830.1 Abelson helper integration site Abelson helper integration site 1 AHR NM_001621.3 aryl hydrocarbon receptor AHSA2;AHA1, activator of heat shock 90kDa protein ATPase homolog 2 AHSA2 NM_152392.1 (yeast),activator of HSP90 ATPase homolog 2 activator of HSP90 ATPase homolog 2, pseudogene APECED;autoimmune regulator (autoimmune polyendocrinopathy candidiasis AIRE NM_000383.2 ectodermal dystrophy) autoimmune regulator AMIGO3 NM_198722.2 adhesion molecule with Ig like domain 3 ANKRD55 NM_024669.2 ankyrin repeat domain 55 ANTXR2 NM_058172.5 anthrax toxin receptor -

SUPPLEMENTARY DATA Supplementary Table 1. Top Ten
SUPPLEMENTARY DATA Supplementary Table 1. Top ten most highly expressed protein-coding genes in the EndoC-βH1 cell line. Expression levels provided for non-mitochondrial genes in EndoC-βH1 and the corresponding expression levels in sorted primary human β-cells (1). Ensembl gene ID Gene Name EndoC-βH1 [RPKM] Primary β cells [RPKM] ENSG00000254647.2 INS 8012.452 166347.111 ENSG00000087086.9 FTL 3090.7454 2066.464 ENSG00000100604.8 CHGA 2853.107 1113.162 ENSG00000099194.5 SCD 1411.631 238.714 ENSG00000118271.5 TTR 1312.8928 1488.996 ENSG00000184009.5 ACTG1 1108.0277 839.681 ENSG00000124172.5 ATP5E 863.42334 254.779 ENSG00000156508.13 EEF1A1 831.17316 637.281 ENSG00000112972.10 HMGCS1 719.7504 22.104 ENSG00000167552.9 TUBA1A 689.1415 511.699 ©2016 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-0361/-/DC1 SUPPLEMENTARY DATA Supplementary Table 2. List of genes selected for inclusion in the primary screen. Expression levels in EndoC-βH1 and sorted primary human β-cells are shown for all genes targeted for silencing in the primary screen, ordered by locus association (1). For gene selection, the following criteria were applied: we first considered (1) all protein-coding genes within 1 Mb of a type 2 diabetes association signal that (2) had non-zero expression (RPKM > 0) in both EndoC-βH1 and primary human β-cells. Previous studies have shown cis-eQTLs to form a relatively tight, symmetrical distribution around the target-gene transcription start site, and a 1 Mb cut-off is thus likely to capture most effector transcripts subject to cis regulation (2-5). -

Supplementary Material Computational Prediction of SARS
Supplementary_Material Computational prediction of SARS-CoV-2 encoded miRNAs and their putative host targets Sheet_1 List of potential stem-loop structures in SARS-CoV-2 genome as predicted by VMir. Rank Name Start Apex Size Score Window Count (Absolute) Direct Orientation 1 MD13 2801 2864 125 243.8 61 2 MD62 11234 11286 101 211.4 49 4 MD136 27666 27721 104 205.6 119 5 MD108 21131 21184 110 204.7 210 9 MD132 26743 26801 119 188.9 252 19 MD56 9797 9858 128 179.1 59 26 MD139 28196 28233 72 170.4 133 28 MD16 2934 2974 76 169.9 71 43 MD103 20002 20042 80 159.3 403 46 MD6 1489 1531 86 156.7 171 51 MD17 2981 3047 131 152.8 38 87 MD4 651 692 75 140.3 46 95 MD7 1810 1872 121 137.4 58 116 MD140 28217 28252 72 133.8 62 122 MD55 9712 9758 96 132.5 49 135 MD70 13171 13219 93 130.2 131 164 MD95 18782 18820 79 124.7 184 173 MD121 24086 24135 99 123.1 45 176 MD96 19046 19086 75 123.1 179 196 MD19 3197 3236 76 120.4 49 200 MD86 17048 17083 73 119.8 428 223 MD75 14534 14600 137 117 51 228 MD50 8824 8870 94 115.8 79 234 MD129 25598 25642 89 115.6 354 Reverse Orientation 6 MR61 19088 19132 88 197.8 271 10 MR72 23563 23636 148 188.8 286 11 MR11 3775 3844 136 185.1 116 12 MR94 29532 29582 94 184.6 271 15 MR43 14973 15028 109 183.9 226 27 MR14 4160 4206 89 170 241 34 MR35 11734 11792 111 164.2 37 52 MR5 1603 1652 89 152.7 118 53 MR57 18089 18132 101 152.7 139 94 MR8 2804 2864 122 137.4 38 107 MR58 18474 18508 72 134.9 237 117 MR16 4506 4540 72 133.8 311 120 MR34 10010 10048 82 132.7 245 133 MR7 2534 2578 90 130.4 75 146 MR79 24766 24808 75 127.9 59 150 MR65 21528 21576 99 127.4 83 180 MR60 19016 19049 70 122.5 72 187 MR51 16450 16482 75 121 363 190 MR80 25687 25734 96 120.6 75 198 MR64 21507 21544 70 120.3 35 206 MR41 14500 14542 84 119.2 94 218 MR84 26840 26894 108 117.6 94 Sheet_2 List of stable stem-loop structures based on MFE.