Supplementary Figure
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Structural Characterization of Helicobacter Pylori Proteins Contributing to Stomach Colonization
Università degli Studi di Padova Dipartimento di Biologia Scuola di Dottorato di Ricerca in Bioscienze e Biotecnologie Indirizzo: Biotecnologie Ciclo XXVIII STRUCTURAL CHARACTERIZATION OF HELICOBACTER PYLORI PROTEINS CONTRIBUTING TO STOMACH COLONIZATION Direttore della Scuola: Ch.mo Prof. Paolo Bernardi Coordinatore di Indirizzo: Ch.ma Prof.ssa Fiorella Lo Schiavo Supervisore: Ch.mo Prof. Giuseppe Zanotti Dottorando: Maria Elena Compostella 31 Gennaio 2016 Università degli Studi di Padova Department of Biology School of Biosciences and Biotechnology Curriculum: Biotechnology XXVIII Cycle STRUCTURAL CHARACTERIZATION OF HELICOBACTER PYLORI PROTEINS CONTRIBUTING TO STOMACH COLONIZATION Director of the Ph.D. School: Ch.mo Prof. Paolo Bernardi Coordinator of the Curriculum: Ch.ma Prof.ssa Fiorella Lo Schiavo Supervisor: Ch.mo Prof. Giuseppe Zanotti Ph.D. Candidate: Maria Elena Compostella 31 January 2016 Contents ABBREVIATIONS AND SYMBOLS IV SUMMARY 9 SOMMARIO 15 1. INTRODUCTION 21 1.1 HELICOBACTER PYLORI 23 1.2 GENETIC VARIABILITY 26 1.2.1 GENOME COMPARISON 26 1.2.1.1 HELICOBACTER PYLORI 26695 26 1.2.1.2 HELICOBACTER PYLORI J99 28 1.2.2 CORE GENOME 30 1.2.3 MECHANISMS GENERATING GENETIC VARIABILITY 31 1.2.3.1 MUTAGENESIS 32 1.2.3.2 RECOMBINATION 35 1.2.4 HELICOBACTER PYLORI AS A “QUASI SPECIES” 37 1.2.5 CLASSIFICATION OF HELICOBACTER PYLORI STRAINS 38 1.3 EPIDEMIOLOGY 40 1.3.1 INCIDENCE AND PREVALENCE OF HELICOBACTER PYLORI INFECTION 40 1.3.2 SOURCE AND TRANSMISSION 42 1.4 ADAPTATION AND GASTRIC COLONIZATION 47 1.4.1 ACID ADAPTATION 49 1.4.2 MOTILITY AND CHEMIOTAXIS 60 1.4.3 ADHESION 65 1.5 PATHOGENESIS AND VIRULENCE FACTORS 72 1.5.1 VACUOLATING CYTOTOXIN A 78 1.5.2 CAG PATHOGENICITY ISLAND AND CYTOTOXIN-ASSOCIATED GENE A 83 1.5.3 NEUTROPHIL-ACTIVATING PROTEIN 90 1.6 HELICOBACTER PYLORI AND GASTRODUODENAL DISEASES 92 1.7 ERADICATION AND POTENTIAL BENEFITS 97 2. -

The Genomes of Polyextremophilic Cyanidiales Contain 1% 2 Horizontally Transferred Genes with Diverse Adaptive Functions 3 4 Alessandro W
bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 The genomes of polyextremophilic Cyanidiales contain 1% 2 horizontally transferred genes with diverse adaptive functions 3 4 Alessandro W. Rossoni1#, Dana C. Price2, Mark Seger3, Dagmar Lyska1, Peter Lammers3, 5 Debashish Bhattacharya4 & Andreas P.M. Weber1* 6 7 1Institute of Plant Biochemistry, Cluster of Excellence on Plant Sciences (CEPLAS), Heinrich 8 Heine University, Universitätsstraße 1, 40225 Düsseldorf, Germany 9 2Department of Plant Biology, Rutgers University, New Brunswick, NJ 08901, USA 10 3Arizona Center for Algae Technology and Innovation, Arizona State University, Mesa, AZ 11 85212, USA 12 4Department of Biochemistry and Microbiology, Rutgers University, New Brunswick, NJ 13 08901, USA 14 15 *Corresponding author: Prof. Dr. Andreas P.M. Weber, 16 e-mail: [email protected] 17 bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 18 Abstract 19 The role and extent of horizontal gene transfer (HGT) in eukaryotes are hotly disputed topics 20 that impact our understanding regarding the origin of metabolic processes and the role of 21 organelles in cellular evolution. -

The Hypercomplex Genome of an Insect Reproductive Parasite Highlights the Importance of Lateral Gene Transfer in Symbiont Biology
This is a repository copy of The hypercomplex genome of an insect reproductive parasite highlights the importance of lateral gene transfer in symbiont biology. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/164886/ Version: Published Version Article: Frost, C.L., Siozios, S., Nadal-Jimenez, P. et al. (4 more authors) (2020) The hypercomplex genome of an insect reproductive parasite highlights the importance of lateral gene transfer in symbiont biology. mBio, 11 (2). e02590-19. ISSN 2150-7511 https://doi.org/10.1128/mbio.02590-19 Reuse This article is distributed under the terms of the Creative Commons Attribution (CC BY) licence. This licence allows you to distribute, remix, tweak, and build upon the work, even commercially, as long as you credit the authors for the original work. More information and the full terms of the licence here: https://creativecommons.org/licenses/ Takedown If you consider content in White Rose Research Online to be in breach of UK law, please notify us by emailing [email protected] including the URL of the record and the reason for the withdrawal request. [email protected] https://eprints.whiterose.ac.uk/ OBSERVATION Ecological and Evolutionary Science crossm The Hypercomplex Genome of an Insect Reproductive Parasite Highlights the Importance of Lateral Gene Transfer in Symbiont Biology Crystal L. Frost,a Stefanos Siozios,a Pol Nadal-Jimenez,a Michael A. Brockhurst,b Kayla C. King,c Alistair C. Darby,a Gregory D. D. Hursta aInstitute of Integrative Biology, University of Liverpool, Liverpool, United Kingdom bDepartment of Animal and Plant Sciences, University of Sheffield, Sheffield, United Kingdom cDepartment of Zoology, University of Oxford, Oxford, United Kingdom Crystal L. -

Copyright by Jeremy Daniel O'connell 2012
Copyright by Jeremy Daniel O’Connell 2012 The Dissertation Committee for Jeremy Daniel O’Connell Certifies that this is the approved version of the following dissertation: Systemic Protein Aggregation in Stress and Aging Restructures Cytoplasmic Architecture Committee: Edward Marcotte, Supervisor Dean Appling Andrew Ellington Makkuni Jayaram Scott Stevens Systemic Protein Aggregation in Stress and Aging Restructures Cytoplasmic Architecture by Jeremy Daniel O’Connell, B.S. Dissertation Presented to the Faculty of the Graduate School of The University of Texas at Austin in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy The University of Texas at Austin September 2012 Dedication Cytisus laburnum, simul vincet omnem To my dad and mom who encouraged and enabled my education with countless sacrifices, I promised this graduation would be the one we would attend, and I am truly sorry I was not swift enough to make that possible. Acknowledgements Foremost, I thank my advisor Edward Marcotte, for not just a second lease on a life in science but one in an amazing lab environment. His intellectual rigor, enduring patience, amazing work ethic, and enthusiasm for discovery were an inspiration. I thank my collaborators in this project: Gwen Stovall, Alice Zhao, Gabe Wu, and Mark Tsechansky for their comradery and support on this great adventure. I thank the talented undergraduates: Maguerite West-Driga, Ariel Royall, and Tyler McDonald who stuck with me. Each of you will soon be a better scientist than I ever will, and I hope you enjoyed and learned from our research together nearly as much as I did. -

The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (Cad) by Phosphorylation and Protein-Protein Interactions
THE REGULATION OF CARBAMOYL PHOSPHATE SYNTHETASE-ASPARTATE TRANSCARBAMOYLASE-DIHYDROOROTASE (CAD) BY PHOSPHORYLATION AND PROTEIN-PROTEIN INTERACTIONS Eric M. Wauson A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Pharmacology. Chapel Hill 2007 Approved by: Lee M. Graves, Ph.D. T. Kendall Harden, Ph.D. Gary L. Johnson, Ph.D. Aziz Sancar M.D., Ph.D. Beverly S. Mitchell, M.D. 2007 Eric M. Wauson ALL RIGHTS RESERVED ii ABSTRACT Eric M. Wauson: The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (CAD) by Phosphorylation and Protein-Protein Interactions (Under the direction of Lee M. Graves, Ph.D.) Pyrimidines have many important roles in cellular physiology, as they are used in the formation of DNA, RNA, phospholipids, and pyrimidine sugars. The first rate- limiting step in the de novo pyrimidine synthesis pathway is catalyzed by the carbamoyl phosphate synthetase II (CPSase II) part of the multienzymatic complex Carbamoyl phosphate synthetase, Aspartate transcarbamoylase, Dihydroorotase (CAD). CAD gene induction is highly correlated to cell proliferation. Additionally, CAD is allosterically inhibited or activated by uridine triphosphate (UTP) or phosphoribosyl pyrophosphate (PRPP), respectively. The phosphorylation of CAD by PKA and ERK has been reported to modulate the response of CAD to allosteric modulators. While there has been much speculation on the identity of CAD phosphorylation sites, no definitive identification of in vivo CAD phosphorylation sites has been performed. Therefore, we sought to determine the specific CAD residues phosphorylated by ERK and PKA in intact cells. -

A Focus on Protein Glycosylation in Lactobacillus
International Journal of Molecular Sciences Review How Sweet Are Our Gut Beneficial Bacteria? A Focus on Protein Glycosylation in Lactobacillus Dimitrios Latousakis and Nathalie Juge * Quadram Institute Bioscience, The Gut Health and Food Safety Institute Strategic Programme, Norwich Research Park, Norwich NR4 7UA, UK; [email protected] * Correspondence: [email protected]; Tel.: +44-(0)-160-325-5068; Fax: +44-(0)-160-350-7723 Received: 22 November 2017; Accepted: 27 December 2017; Published: 3 January 2018 Abstract: Protein glycosylation is emerging as an important feature in bacteria. Protein glycosylation systems have been reported and studied in many pathogenic bacteria, revealing an important diversity of glycan structures and pathways within and between bacterial species. These systems play key roles in virulence and pathogenicity. More recently, a large number of bacterial proteins have been found to be glycosylated in gut commensal bacteria. We present an overview of bacterial protein glycosylation systems (O- and N-glycosylation) in bacteria, with a focus on glycoproteins from gut commensal bacteria, particularly Lactobacilli. These emerging studies underscore the importance of bacterial protein glycosylation in the interaction of the gut microbiota with the host. Keywords: protein glycosylation; gut commensal bacteria; Lactobacillus; glycoproteins; adhesins; lectins; O-glycosylation; N-glycosylation; probiotics 1. Introduction Protein glycosylation, i.e., the covalent attachment of a carbohydrate moiety onto a protein, is a highly ubiquitous protein modification in nature, and considered to be one of the post-translational modifications (PTM) targeting the most diverse group of proteins [1]. Although it was originally believed to be restricted to eukaryotic systems and later to archaea, it has become apparent nowadays that protein glycosylation is a common feature in all three domains of life. -
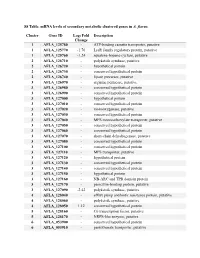
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

In Silico Evolutionary Analysis of Helicobacter Pylori Outer Membrane Phospholipase a (OMPLA) Hilde S Vollan1*, Tone Tannæs1, Yoshio Yamaoka2 and Geir Bukholm3,4
Vollan et al. BMC Microbiology 2012, 12:206 http://www.biomedcentral.com/1471-2180/12/206 RESEARCH ARTICLE Open Access In silico evolutionary analysis of Helicobacter pylori outer membrane phospholipase A (OMPLA) Hilde S Vollan1*, Tone Tannæs1, Yoshio Yamaoka2 and Geir Bukholm3,4 Abstract Background: In the past decade, researchers have proposed that the pldA gene for outer membrane phospholipase A (OMPLA) is important for bacterial colonization of the human gastric ventricle. Several conserved Helicobacter pylori genes have distinct genotypes in different parts of the world, biogeographic patterns that can be analyzed through phylogenetic trees. The current study will shed light on the importance of the pldA gene in H. pylori. In silico sequence analysis will be used to investigate whether the bacteria are in the process of preserving, optimizing, or rejecting the pldA gene. The pldA gene will be phylogenetically compared to other housekeeping (HK) genes, and a possible origin via horizontal gene transfer (HGT) will be evaluated through both intra- and inter- species evolutionary analyses. Results: In this study, pldA gene sequences were phylogenetically analyzed and compared with a large reference set of concatenated HK gene sequences. A total of 246 pldA nucleotide sequences were used; 207 were from Norwegian isolates, 20 were from Korean isolates, and 19 were from the NCBI database. Best-fit evolutionary models were determined with MEGA5 ModelTest for the pldA (K80 + I + G) and HK (GTR + I + G) sequences, and maximum likelihood trees were constructed. Both HK and pldA genes showed biogeographic clustering. Horizontal gene transfer was inferred based on significantly different GC contents, the codon adaptation index, and a phylogenetic conflict between a tree of OMPLA protein sequences representing 171 species and a tree of the AtpA HK protein for 169 species. -

Transitions in Symbiosis: Evidence for Environmental Acquisition and Social Transmission Within a Clade of Heritable Symbionts
The ISME Journal (2021) 15:2956–2968 https://doi.org/10.1038/s41396-021-00977-z ARTICLE Transitions in symbiosis: evidence for environmental acquisition and social transmission within a clade of heritable symbionts 1,2 3 2 4 2 Georgia C. Drew ● Giles E. Budge ● Crystal L. Frost ● Peter Neumann ● Stefanos Siozios ● 4 2 Orlando Yañez ● Gregory D. D. Hurst Received: 5 August 2020 / Revised: 17 March 2021 / Accepted: 6 April 2021 / Published online: 3 May 2021 © The Author(s) 2021. This article is published with open access Abstract A dynamic continuum exists from free-living environmental microbes to strict host-associated symbionts that are vertically inherited. However, knowledge of the forces that drive transitions in symbiotic lifestyle and transmission mode is lacking. Arsenophonus is a diverse clade of bacterial symbionts, comprising reproductive parasites to coevolving obligate mutualists, in which the predominant mode of transmission is vertical. We describe a symbiosis between a member of the genus Arsenophonus and the Western honey bee. The symbiont shares common genomic and predicted metabolic properties with the male-killing symbiont Arsenophonus nasoniae, however we present multiple lines of evidence that the bee 1234567890();,: 1234567890();,: Arsenophonus deviates from a heritable model of transmission. Field sampling uncovered spatial and seasonal dynamics in symbiont prevalence, and rapid infection loss events were observed in field colonies and laboratory individuals. Fluorescent in situ hybridisation showed Arsenophonus localised in the gut, and detection was rare in screens of early honey bee life stages. We directly show horizontal transmission of Arsenophonus between bees under varying social conditions. We conclude that honey bees acquire Arsenophonus through a combination of environmental exposure and social contacts. -

Mrna Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection
International Journal of Molecular Sciences Review mRNA Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection 1, 1, 2 1,3, Shuqin Xu y, Kunpeng Yang y, Rose Li and Lu Zhang * 1 State Key Laboratory of Genetic Engineering, Institute of Genetics, School of Life Science, Fudan University, Shanghai 200438, China; [email protected] (S.X.); [email protected] (K.Y.) 2 M.B.B.S., School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; [email protected] 3 Shanghai Engineering Research Center of Industrial Microorganisms, Shanghai 200438, China * Correspondence: [email protected]; Tel.: +86-13524278762 These authors contributed equally to this work. y Received: 30 July 2020; Accepted: 30 August 2020; Published: 9 September 2020 Abstract: Messenger ribonucleic acid (mRNA)-based drugs, notably mRNA vaccines, have been widely proven as a promising treatment strategy in immune therapeutics. The extraordinary advantages associated with mRNA vaccines, including their high efficacy, a relatively low severity of side effects, and low attainment costs, have enabled them to become prevalent in pre-clinical and clinical trials against various infectious diseases and cancers. Recent technological advancements have alleviated some issues that hinder mRNA vaccine development, such as low efficiency that exist in both gene translation and in vivo deliveries. mRNA immunogenicity can also be greatly adjusted as a result of upgraded technologies. In this review, we have summarized details regarding the optimization of mRNA vaccines, and the underlying biological mechanisms of this form of vaccines. Applications of mRNA vaccines in some infectious diseases and cancers are introduced. It also includes our prospections for mRNA vaccine applications in diseases caused by bacterial pathogens, such as tuberculosis. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Metatranscriptomic Analysis of Community Structure And
School of Environmental Sciences Metatranscriptomic analysis of community structure and metabolism of the rhizosphere microbiome by Thomas Richard Turner Submitted in partial fulfilment of the requirement for the degree of Doctor of Philosophy, September 2013 This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that its copyright rests with the author and that use of any information derived there from must be in accordance with current UK Copyright Law. In addition, any quotation or extract must include full attribution. i Declaration I declare that this is an account of my own research and has not been submitted for a degree at any other university. The use of material from other sources has been properly and fully acknowledged, where appropriate. Thomas Richard Turner ii Acknowledgements I would like to thank my supervisors, Phil Poole and Alastair Grant, for their continued support and guidance over the past four years. I’m grateful to all members of my lab, both past and present, for advice and friendship. Graham Hood, I don’t know how we put up with each other, but I don’t think I could have done this without you. Cheers Salt! KK, thank you for all your help in the lab, and for Uma’s biryanis! Andrzej Tkatcz, thanks for the useful discussions about our projects. Alison East, thank you for all your support, particularly ensuring Graham and I did not kill each other. I’m grateful to Allan Downie and Colin Murrell for advice. For sequencing support, I’d like to thank TGAC, particularly Darren Heavens, Sophie Janacek, Kirsten McKlay and Melanie Febrer, as well as John Walshaw, Mark Alston and David Swarbreck for bioinformatic support.