Fossils Matter: Improved Estimates of Divergence Times in Pinus Reveal
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

(Pinus Taeda L.) with Related Species
RESEARCH ARTICLE Complete chloroplast genome sequence and comparative analysis of loblolly pine (Pinus taeda L.) with related species Sajjad Asaf1, Abdul Latif Khan1, Muhammad Aaqil Khan2, Raheem Shahzad2, Lubna3, Sang Mo Kang2, Ahmed Al-Harrasi1, Ahmed Al-Rawahi1, In-Jung Lee2,4* 1 Chair of Oman's Medicinal Plants & Marine Natural Products, University of Nizwa, Nizwa, Oman, 2 School of Applied Biosciences, Kyungpook National University, Daegu, Republic of Korea, 3 Department of Botany, Garden Campus, Abdul Wali Khan University Mardan, Mardan, Pakistan, 4 Research Institute for Dok-do and Ulleung-do Island, Kyungpook National University, Daegu, Republic of Korea a1111111111 a1111111111 * [email protected] a1111111111 a1111111111 a1111111111 Abstract Pinaceae, the largest family of conifers, has a diversified organization of chloroplast (cp) genomes with two typical highly reduced inverted repeats (IRs). In the current study, we determined the complete sequence of the cp genome of an economically and ecologically OPEN ACCESS important conifer tree, the loblolly pine (Pinus taeda L.), using Illumina paired-end sequenc- Citation: Asaf S, Khan AL, Khan MA, Shahzad R, ing and compared the sequence with those of other pine species. The results revealed a Lubna , Kang SM, et al. (2018) Complete chloroplast genome sequence and comparative genome size of 121,531 base pairs (bp) containing a pair of 830-bp IR regions, distinguished analysis of loblolly pine (Pinus taeda L.) with by a small single copy (42,258 bp) and large single copy (77,614 bp) region. The chloroplast related species. PLoS ONE 13(3): e0192966. genome of P. taeda encodes 120 genes, comprising 81 protein-coding genes, four ribo- https://doi.org/10.1371/journal.pone.0192966 somal RNA genes, and 35 tRNA genes, with 151 randomly distributed microsatellites. -

A New Classification of Marginal Resin Ducts Improves Understanding of Hard Pine (Pinaceae) Diversity in Taiwan
Flora 209 (2014) 414–425 Contents lists available at ScienceDirect Flora journal homepage: www.elsevier.com/locate/flora A new classification of marginal resin ducts improves understanding of hard pine (Pinaceae) diversity in Taiwan a,b, a,c d d Chiou-Rong Sheue *, Hsiu-Chin Chang , Yuen-Po Yang , Ho-Yih Liu , a,b,e c Peter Chesson , Fu-Hsiung Hsu a Department of Life Sciences, National Chung Hsing University, 250 Kuo Kuang Road., Taichung 402, Taiwan b Center of Global Change Biology, National Chung Hsing University, 250, Kuo Kuang Road, Taichung 402, Taiwan c Department of Biological Resources, National Chiayi University, 300 Syuefu Road., Chiayi City 600, Taiwan d Department of Biological Sciences, National Sun Yat-sen University, Kaohsiung 804, Taiwan e Department of Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ 85721, USA A R T I C L E I N F O A B S T R A C T Article history: Resin ducts provide important characters for classifying the Pinaceae. Here we study Pinus massoniana Received 18 November 2013 and P. taiwanensis and show that the generally-used term marginal (=external) resin duct, applied to Accepted 5 June 2014 ducts in needle leaves, needs to be further differentiated into marginal (strongly attaching to the dermal Edited by R. Lösch tissue, and lacking the complete ring structure formed by the sheath cells) and submarginal ducts Available online 28 June 2014 (adjacent to hypodermal cells, with a complete ring structure formed by the sheath cells). On this basis P. massoniana and P. taiwanensis, which are nearly indistinguishable based on external morphology, are Keywords: clearly differentiated. -

Climate Change Vulnerability and Adaptation in the Intermountain Region Part 1
United States Department of Agriculture Climate Change Vulnerability and Adaptation in the Intermountain Region Part 1 Forest Rocky Mountain General Technical Report Service Research Station RMRS-GTR-375 April 2018 Halofsky, Jessica E.; Peterson, David L.; Ho, Joanne J.; Little, Natalie, J.; Joyce, Linda A., eds. 2018. Climate change vulnerability and adaptation in the Intermountain Region. Gen. Tech. Rep. RMRS-GTR-375. Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station. Part 1. pp. 1–197. Abstract The Intermountain Adaptation Partnership (IAP) identified climate change issues relevant to resource management on Federal lands in Nevada, Utah, southern Idaho, eastern California, and western Wyoming, and developed solutions intended to minimize negative effects of climate change and facilitate transition of diverse ecosystems to a warmer climate. U.S. Department of Agriculture Forest Service scientists, Federal resource managers, and stakeholders collaborated over a 2-year period to conduct a state-of-science climate change vulnerability assessment and develop adaptation options for Federal lands. The vulnerability assessment emphasized key resource areas— water, fisheries, vegetation and disturbance, wildlife, recreation, infrastructure, cultural heritage, and ecosystem services—regarded as the most important for ecosystems and human communities. The earliest and most profound effects of climate change are expected for water resources, the result of declining snowpacks causing higher peak winter -

Non-Wood Forest Products from Conifers
Page 1 of 8 NON -WOOD FOREST PRODUCTS 12 Non-Wood Forest Products From Conifers FAO - Food and Agriculture Organization of the United Nations The designations employed and the presentation of material in this publication do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. M-37 ISBN 92-5-104212-8 (c) FAO 1995 TABLE OF CONTENTS FOREWORD ACKNOWLEDGMENTS ABBREVIATIONS INTRODUCTION CHAPTER 1 - AN OVERVIEW OF THE CONIFERS WHAT ARE CONIFERS? DISTRIBUTION AND ABUNDANCE USES CHAPTER 2 - CONIFERS IN HUMAN CULTURE FOLKLORE AND MYTHOLOGY RELIGION POLITICAL SYMBOLS ART CHAPTER 3 - WHOLE TREES LANDSCAPE AND ORNAMENTAL TREES Page 2 of 8 Historical aspects Benefits Species Uses Foliage effect Specimen and character trees Shelter, screening and backcloth plantings Hedges CHRISTMAS TREES Historical aspects Species Abies spp Picea spp Pinus spp Pseudotsuga menziesii Other species Production and trade BONSAI Historical aspects Bonsai as an art form Bonsai cultivation Species Current status TOPIARY CONIFERS AS HOUSE PLANTS CHAPTER 4 - FOLIAGE EVERGREEN BOUGHS Uses Species Harvesting, management and trade PINE NEEDLES Mulch Decorative baskets OTHER USES OF CONIFER FOLIAGE CHAPTER 5 - BARK AND ROOTS TRADITIONAL USES Inner bark as food Medicinal uses Natural dyes Other uses TAXOL Description and uses Harvesting methods Alternative -

Phytosociological Analysis of Pine Forest at Indus Kohistan, Kpk, Pakistan
Pak. J. Bot., 48(2): 575-580, 2016. PHYTOSOCIOLOGICAL ANALYSIS OF PINE FOREST AT INDUS KOHISTAN, KPK, PAKISTAN ADAM KHAN1, MOINUDDIN AHMED2, MUHAMMAD FAHEEM SIDDIQUI*3, JAVED IQBAL1 AND MUHAMMAD WAHAB4 1Laboratory of plant ecology and Dendrochronology, Department of Botany, Federal Urdu University, Gulshan-e-Iqbal Campus Karachi, Pakistan 2Department of Earth and Environmental Systems, 600 Chestnut Street Indiana State University, Terre Haute, IN, USA 3Department of Botany, University of Karachi, Karachi-75270, Pakistan 4Institute of Botany, Chinese Academy of Sciences, Beijing, China *Corresponding author’s email: [email protected] Abstract The study was carried out to describe the pine communities at Indus Kohistan valley in quantitative term. Thirty stands of relatively undisturbed vegetation were selected for sampling. Quantitative sampling was carried out by Point Centered Quarter (PCQ) method. Seven tree species were common in the Indus Kohistan valley. Cedrus deodara was recorded from twenty eight different locations and exhibited the highest mean importance value while Pinus wallichiana was recorded from 23 different locations and exhibited second highest mean importance value. Third most occurring species was Abies pindrow that attained the third highest mean importance value and Picea smithiana was recorded from eight different locations and attained fourth highest importance value while it was first dominant in one stand and second dominant in four stands. Pinus gerardiana, Quercus baloot and Taxus fuana were the rare species in this area, these species attained low mean importance value. Six communities and four monospecific stands of Cedrus deodara were recognized. Cedrus-Pinus community was the most occurring community, which was recorded from 13 different stands. -
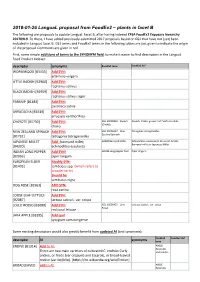
2018-01-26 Langual Proposal from Foodex2 – Plants in Facet B
2018-01-26 LanguaL proposal from FoodEx2 – plants in facet B The following are proposals to update LanguaL Facet B, after having indexed EFSA FoodEx2 Exposure hierarchy 20170919. To these, I have added previously-submitted 2017 proposals based on GS1 that have not (yet) been included in LanguaL facet B. GS1 terms and FoodEx2 terms in the following tables are just given to indicate the origin of the proposal. Comments are given in red. First, some simple additions of terms to the SYNONYM field, to make it easier to find descriptors in the LanguaL Food Product Indexer: descriptor synonyms FoodEx2 term FoodEx2 def WORMWOOD [B3433] Add SYN: artemisia vulgaris LITTLE RADISH [B2960] Add SYN: raphanus sativus BLACK RADISH [B2959] Add SYN: raphanus sativus niger PARSNIP [B1483] Add SYN: pastinaca sativa ARRACACHA [B3439] Add SYN: arracacia xanthorrhiza CHAYOTE [B1730] Add SYN: GS1 10006356 - Squash Squash, Choko, grown from Sechium edule (Choko) choko NEW ZEALAND SPINACH Add SYN: GS1 10006427 - New- Tetragonia tetragonoides Zealand Spinach [B1732] tetragonia tetragonoides JAPANESE MILLET Add : barnyard millet; A000Z Barnyard millet Echinochloa esculenta (A. Braun) H. Scholz, Barnyard millet or Japanese Millet. [B4320] echinochloa esculenta INDIAN LONG PEPPER Add SYN! A019B Long pepper fruit Piper longum [B2956] piper longum EUROPEAN ELDER Modify SYN: [B1403] sambucus spp. (which refers to broader term) Should be sambucus nigra DOG ROSE [B2961] ADD SYN: rosa canina LOOSE LEAF LETTUCE Add SYN: [B2087] lactusa sativa L. var. crispa LOLLO ROSSO [B2088] Add SYN: GS1 10006425 - Lollo Lactuca sativa L. var. crispa Rosso red coral lettuce JAVA APPLE [B3395] Add syn! syzygium samarangense Some existing descriptors would also greatly benefit from updated AI (and synonyms): FoodEx2 FoodEx2 def descriptor AI synonyms term ENDIVE [B1314] Add to AI: A00LD Escaroles There are two main varieties of cultivated C. -

Biodiversity Conservation in Botanical Gardens
AgroSMART 2019 International scientific and practical conference ``AgroSMART - Smart solutions for agriculture'' Volume 2019 Conference Paper Biodiversity Conservation in Botanical Gardens: The Collection of Pinaceae Representatives in the Greenhouses of Peter the Great Botanical Garden (BIN RAN) E M Arnautova and M A Yaroslavceva Department of Botanical garden, BIN RAN, Saint-Petersburg, Russia Abstract The work researches the role of botanical gardens in biodiversity conservation. It cites the total number of rare and endangered plants in the greenhouse collection of Peter the Great Botanical garden (BIN RAN). The greenhouse collection of Pinaceae representatives has been analysed, provided with a short description of family, genus and certain species, presented in the collection. The article highlights the importance of Pinaceae for various industries, decorative value of plants of this group, the worth of the pinaceous as having environment-improving properties. In Corresponding Author: the greenhouses there are 37 species of Pinaceae, of 7 geni, all species have a E M Arnautova conservation status: CR -- 2 species, EN -- 3 species, VU- 3 species, NT -- 4 species, LC [email protected] -- 25 species. For most species it is indicated what causes depletion. Most often it is Received: 25 October 2019 the destruction of natural habitats, uncontrolled clearance, insect invasion and diseases. Accepted: 15 November 2019 Published: 25 November 2019 Keywords: biodiversity, botanical gardens, collections of tropical and subtropical plants, Pinaceae plants, conservation status Publishing services provided by Knowledge E E M Arnautova and M A Yaroslavceva. This article is distributed under the terms of the Creative Commons 1. Introduction Attribution License, which permits unrestricted use and Nowadays research of biodiversity is believed to be one of the overarching goals for redistribution provided that the original author and source are the modern world. -

Texas Big Tree Registry a List of the Largest Trees in Texas Sponsored by Texas a & M Forest Service
Texas Big Tree Registry A list of the largest trees in Texas Sponsored by Texas A & M Forest Service Native and Naturalized Species of Texas: 320 ( D indicates species naturalized to Texas) Common Name (also known as) Latin Name Remarks Cir. Threshold acacia, Berlandier (guajillo) Senegalia berlandieri Considered a shrub by B. Simpson 18'' or 1.5 ' acacia, blackbrush Vachellia rigidula Considered a shrub by Simpson 12'' or 1.0 ' acacia, Gregg (catclaw acacia, Gregg catclaw) Senegalia greggii var. greggii Was named A. greggii 55'' or 4.6 ' acacia, Roemer (roundflower catclaw) Senegalia roemeriana 18'' or 1.5 ' acacia, sweet (huisache) Vachellia farnesiana 100'' or 8.3 ' acacia, twisted (huisachillo) Vachellia bravoensis Was named 'A. tortuosa' 9'' or 0.8 ' acacia, Wright (Wright catclaw) Senegalia greggii var. wrightii Was named 'A. wrightii' 70'' or 5.8 ' D ailanthus (tree-of-heaven) Ailanthus altissima 120'' or 10.0 ' alder, hazel Alnus serrulata 18'' or 1.5 ' allthorn (crown-of-thorns) Koeberlinia spinosa Considered a shrub by Simpson 18'' or 1.5 ' anacahuita (anacahuite, Mexican olive) Cordia boissieri 60'' or 5.0 ' anacua (anaqua, knockaway) Ehretia anacua 120'' or 10.0 ' ash, Carolina Fraxinus caroliniana 90'' or 7.5 ' ash, Chihuahuan Fraxinus papillosa 12'' or 1.0 ' ash, fragrant Fraxinus cuspidata 18'' or 1.5 ' ash, green Fraxinus pennsylvanica 120'' or 10.0 ' ash, Gregg (littleleaf ash) Fraxinus greggii 12'' or 1.0 ' ash, Mexican (Berlandier ash) Fraxinus berlandieriana Was named 'F. berlandierana' 120'' or 10.0 ' ash, Texas Fraxinus texensis 60'' or 5.0 ' ash, velvet (Arizona ash) Fraxinus velutina 120'' or 10.0 ' ash, white Fraxinus americana 100'' or 8.3 ' aspen, quaking Populus tremuloides 25'' or 2.1 ' baccharis, eastern (groundseltree) Baccharis halimifolia Considered a shrub by Simpson 12'' or 1.0 ' baldcypress (bald cypress) Taxodium distichum Was named 'T. -

Bunzo Hayata and His Contributions to the Flora of Taiwan
TAIWANIA, 54(1): 1-27, 2009 INVITED PAPER Bunzo Hayata and His Contributions to the Flora of Taiwan Hiroyoshi Ohashi Botanical Garden, Tohoku University, Sendai 980-0962, Japan. Email: [email protected] (Manuscript received 10 September 2008; accepted 24 October 2008) ABSTRACT: Bunzo Hayata was the founding father of the study of the flora of Taiwan. From 1900 to 1921 Taiwan’s flora was the focus of his attention. During that time he named about 1600 new taxa of vascular plants from Taiwan. Three topics are presented in this paper: a biography of Bunzo Hayata; Hayata’s contributions to the flora of Taiwan; and the current status of Hayata’s new taxa. The second item includes five subitems: i) floristic studies of Taiwan before Hayata, ii) the first 10 years of Hayata’s study of the flora of Taiwan, iii) Taiwania, iv) the second 10 years, and v) Hayata’s works after the flora of Taiwan. The third item is the first step of the evaluation of Hayata’s contribution to the flora of Taiwan. New taxa in Icones Plantarum Formosanarum vol. 10 and the gymnosperms described by Hayata from Taiwan are exampled in this paper. KEY WORDS: biography, Cupressaceae, flora of Taiwan, gymnosperms, Hayata Bunzo, Icones Plantarum Formosanarum, Taiwania, Taxodiaceae. 1944). Wu (1997) wrote a biography of Hayata in INTRODUCTION Chinese as a botanist who worked in Taiwan during the period of Japanese occupation based biographies and Bunzo Hayata (早田文藏) [1874-1934] (Fig. 1) was memoirs written in Japanese. Although there are many a Japanese botanist who described numerous new taxa in articles on the works of Hayata in Japanese, many of nearly every family of vascular plants of Taiwan. -

Disturbances Influence Trait Evolution in Pinus
Master's Thesis Diversify or specialize: Disturbances influence trait evolution in Pinus Supervision by: Prof. Dr. Elena Conti & Dr. Niklaus E. Zimmermann University of Zurich, Institute of Systematic Botany & Swiss Federal Research Institute WSL Birmensdorf Landscape Dynamics Bianca Saladin October 2013 Front page: Forest of Pinus taeda, northern Florida, 1/2013 Table of content 1 STRONG PHYLOGENETIC SIGNAL IN PINE TRAITS 5 1.1 ABSTRACT 5 1.2 INTRODUCTION 5 1.3 MATERIAL AND METHODS 8 1.3.1 PHYLOGENETIC INFERENCE 8 1.3.2 TRAIT DATA 9 1.3.3 PHYLOGENETIC SIGNAL 9 1.4 RESULTS 11 1.4.1 PHYLOGENETIC INFERENCE 11 1.4.2 PHYLOGENETIC SIGNAL 12 1.5 DISCUSSION 14 1.5.1 PHYLOGENETIC INFERENCE 14 1.5.2 PHYLOGENETIC SIGNAL 16 1.6 CONCLUSION 17 1.7 ACKNOWLEDGEMENTS 17 1.8 REFERENCES 19 2 THE ROLE OF FIRE IN TRIGGERING DIVERSIFICATION RATES IN PINE SPECIES 21 2.1 ABSTRACT 21 2.2 INTRODUCTION 21 2.3 MATERIAL AND METHODS 24 2.3.1 PHYLOGENETIC INFERENCE 24 2.3.2 DIVERSIFICATION RATE 24 2.4 RESULTS 25 2.4.1 PHYLOGENETIC INFERENCE 25 2.4.2 DIVERSIFICATION RATE 25 2.5 DISCUSSION 29 2.5.1 DIVERSIFICATION RATE IN RESPONSE TO FIRE ADAPTATIONS 29 2.5.2 DIVERSIFICATION RATE IN RESPONSE TO DISTURBANCE, STRESS AND PLEIOTROPIC COSTS 30 2.5.3 CRITICAL EVALUATION OF THE ANALYSIS PATHWAY 33 2.5.4 PHYLOGENETIC INFERENCE 34 2.6 CONCLUSIONS AND OUTLOOK 34 2.7 ACKNOWLEDGEMENTS 35 2.8 REFERENCES 36 3 SUPPLEMENTARY MATERIAL 39 3.1 S1 - ACCESSION NUMBERS OF GENE SEQUENCES 40 3.2 S2 - TRAIT DATABASE 44 3.3 S3 - SPECIES DISTRIBUTION MAPS 58 3.4 S4 - DISTRIBUTION OF TRAITS OVER PHYLOGENY 81 3.5 S5 - PHYLOGENETIC SIGNAL OF 19 BIOCLIM VARIABLES 84 3.6 S6 – COMPLETE LIST OF REFERENCES 85 2 Introduction to the Master's thesis The aim of my master's thesis was to assess trait and niche evolution in pines within a phylogenetic comparative framework. -

Studies on Drying, Packaging and Storage of Solar Tunnel Dried Chilgoza Nuts
Available online a t www.scholarsresearchlibrary.com Scholars Research Library Archives of Applied Science Research, 2012, 4 (3):1311-1319 (http://scholarsresearchlibrary.com/archive.html) ISSN 0975-508X CODEN (USA) AASRC9 Studies on drying, packaging and storage of solar tunnel dried chilgoza nuts N. S Thakur, Sharma S, Joshi V. K, Thakur K. S and Jindal N Department of Food Science and Technology, Dr YS Parmar University of Horticulture and Forestry, Nauni-Solan, HP ______________________________________________________________________________ ABSTRACT Chilgoza (Pinus gerardiana) is one of the pine nuts among six species found in India which produce edible nuts. Because of the traditional handling of this nut by tribals, it lasts only for few weeks in the market. Studies were undertaken to compare the solar drying modes for drying of this nut and screen out the suitable packaging material for its storage. Extracted nuts were dried under three solar drying means like solar cabinet drier (46-52 ⁰C), solar tunnel drier (43-47 ⁰C) and open sun (18-22 ⁰C). Solar tunnel drier was found to be best drying mode for drying quality nuts as compare to the others. So, nuts dried in this drier were packed in five different packaging materials and stored under ambient conditions for six months. The some physico-chemical quality characteristics like a w (0.208), oil (49.1%) total carbohydrates ( 24.9%), and proteins ( 11.8%) and sensory quality attributes of packed nuts were retained better in glass jars closely followed by aluminum laminate pouch after six months of storage as compared to others. Solar tunnel drier was the best drying mode and glass jar as well as aluminum laminate pouch were the best materials for packaging and storage. -

Landa Park Tree Management Plan
PARKS AND RECREATION DEPARTMENT Landa Park Tree Maintenance Plan CITY OF NEW BRAUNFELS PARKS AND RECREATION DEPARTMENT Mission statement Our mission is to afford diverse opportunities and access for all residents and visitors through innovative programs and facilities, open space preservation and economic enhancement. Vision Statement Our Vision statement is to enhance the well being of our community through laughter, play, conservation and discovery. TABLE OF CONTENTS LANDA PARK TREE MAINTENANCE PLAN EXECUTIVE SUMMARY..………………………..……………..………………………… Page 1 HISTORY ……………………………………………………………………………….…... Page 1 BENEFITS OF TREES………………………………….………………………................Page 2 2015 MANAGEMENT GOALS……………………………………………………………. Page 3 SUMMARY OF MAINTENANCE RECOMMENDATIONS FOR LANDA PARK……... Page 5 LANDA PARK TREE INVENTORY ………………………………………..…………….. Page 8 LANDA PARK FORESTRY STRATEGIES…………………………………………..…. Page 11 Management Zones………………………………………………............................ Page 11 Maintenance Guidelines ………………………………………............................... Page 11 Tree Specific Issues…………………………………………………........................ Page 15 Tree Protection……………………………………………………….….................... Page 16 Tree Planting Plan…………………………………………….………...................... Page 17 Species Diversity and Selection…………………………………............................ Page 17 Stem Density………………………………………………………............................ Page 19 Planting in Specific Locations…………………………………….………………….. Page 21 Outreach……………………………………………………………........................... Page 21 Education………………………………………………………………......................