Der Bahngigant / A-Train 9 Zugliste (Inklusive Fanpatch)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Race 1 1 1 2 2 3 2 4 3 5 3 6 4 7 4 8
TOKYO SUNDAY,MAY 19TH Post Time 10:05 1 ! Race Dirt 1400m THREE−YEAR−OLDS Course Record:31May.08 1:21.9 F&M DES,WEIGHT FOR AGE,MAIDEN Value of race: 9,550,000 Yen 1st 2nd 3rd 4th 5th Added Money(Yen) 5,000,000 2,000,000 1,300,000 750,000 500,000 Stakes Money(Yen) 0 0 0 Ow. Dearest 2,750,000 S 40004 Life60105M 20101 1 54.0 Kosei Miura(6.3%,21−17−29−265,20th) Turf10001 I 00000 1 Dear Aspen(JPN) Dirt50104L 00000 .Captain Steve(0.47) .Forty Niner F3,ch. Yasuhito Tamura(9.6%,14−15−20−97,10th) Course20101E 00000 Wht. /Keiai Patra /Sachi George 20May.10 Nonomiya Bokujo Wet 00000 5May.13 TOKYO MDN D1400St 2 2 1:26.7 4th/15 Kosei Miura 54.0 454# Printemps Bijou 1:25.7 <5> Himesakura <1 1/4> Earth the Three 20Apr.13 TOKYO MDN D1400St 3 3 1:26.9 2nd/16 Kosei Miura 54.0 454% Seiyu Smile 1:26.9 <NK> Dear Aspen <2 1/2> Denko Arrows 24Dec.12 NAKAYAMA MDN D1200Go 2 4 1:13.4 7th/16 Masaki Katsuura 54.0 450+ Hayabusa Pekochan 1:12.8 <NS> Nikiya Dia <1/2> Mary’s Mie 11Nov.12 FUKUSHIMA MDN D1150St 7 5 DQ&P 1:11.5 (6)14th/16 Kyosuke Maruta 54.0 448* Land Queen 1:10.2 <HD> Erimo Feather <1 1/2> Asuka Blanche 29Jul.12 SAPPORO MDN D1000St 8 8 1:01.4 7th/11 Yusuke Fujioka 54.0 436( Regis 0:59.9 <2> Passion <NK> Lapisblau Ow. -

East Japan Railway Company Shin-Hakodate-Hokuto
ANNUAL REPORT 2017 For the year ended March 31, 2017 Pursuing We have been pursuing initiatives in light of the Group Philosophy since 1987. Annual Report 2017 1 Tokyo 1988 2002 We have been pursuing our Eternal Mission while broadening our Unlimited Potential. 1988* 2002 Operating Revenues Operating Revenues ¥1,565.7 ¥2,543.3 billion billion Operating Revenues Operating Income Operating Income Operating Income ¥307.3 ¥316.3 billion billion Transportation (“Railway” in FY1988) 2017 Other Operations (in FY1988) Retail & Services (“Station Space Utilization” in FY2002–2017) Real Estate & Hotels * Fiscal 1988 figures are nonconsolidated. (“Shopping Centers & Office Buildings” in FY2002–2017) Others (in FY2002–2017) Further, other operations include bus services. April 1987 July 1992 March 1997 November 2001 February 2002 March 2004 Establishment of Launch of the Launch of the Akita Launch of Launch of the Station Start of Suica JR East Yamagata Shinkansen Shinkansen Suica Renaissance program with electronic money Tsubasa service Komachi service the opening of atré Ueno service 2 East Japan Railway Company Shin-Hakodate-Hokuto Shin-Aomori 2017 Hachinohe Operating Revenues ¥2,880.8 billion Akita Morioka Operating Income ¥466.3 billion Shinjo Yamagata Sendai Niigata Fukushima Koriyama Joetsumyoko Shinkansen (JR East) Echigo-Yuzawa Conventional Lines (Kanto Area Network) Conventional Lines (Other Network) Toyama Nagano BRT (Bus Rapid Transit) Lines Kanazawa Utsunomiya Shinkansen (Other JR Companies) Takasaki Mito Shinkansen (Under Construction) (As of June 2017) Karuizawa Omiya Tokyo Narita Airport Hachioji Chiba 2017Yokohama Transportation Retail & Services Real Estate & Hotels Others Railway Business, Bus Services, Retail Sales, Restaurant Operations, Shopping Center Operations, IT & Suica business such as the Cleaning Services, Railcar Advertising & Publicity, etc. -

Race 1 1 1 2 2 3 2 4 3 5 3 6 4 7 4 8
HANSHIN SUNDAY,APRIL 7TH Post Time 10:05 1 ! Race Dirt 1800m THREE−YEAR−OLDS Course Record:10Jul.04 1:48.5 DES,WEIGHT FOR AGE,MAIDEN Value of race: 9,550,000 Yen 1st 2nd 3rd 4th 5th Added Money(Yen) 5,000,000 2,000,000 1,300,000 750,000 500,000 Stakes Money(Yen) 0 0 0 Ow. Yoshio Matsumoto 1,450,000 S 00000 Life40004M 30003 1 B 56.0 Ryuji Wada(5.9%,13−29−18−161,22nd) Turf10001 I 00000 1 Meisho Imawaka(JPN) Dirt30003L 10001 0Symboli Kris S(0.97) 0Afleet C3,b. Katsumi Minai(11.7%,9−6−3−59,19th) Course10001E 00000 Wht. 1Prophecy Lights 1Storm the Mint 8Mar.10 Fujiwara Farm Wet 10001 24Mar.13 HANSHIN MDN D1800St 4456 1:55.54th/13Kyosuke Kokubun 56.0 548% Manoir 1:54.9 <1> Zillion <1 3/4> Meiner Gainer 2Mar.13 HANSHIN MDN T2200Go2222 2:18.78th/16RyujiWada 56.0548( Hamano Grade 2:17.6 <1/2> Danon Symphony <3> Sunrise Way 12Jan.13 KYOTO MDN D1800St 11 11 9 13 1:57.3 14th/16 Yutaka Take 56.0 558# Copano Rickey 1:52.4 <5> Dynamic War <1/2> Courage d’Or 16Dec.12 CHUKYO NWC D1800Mu4545 1:57.35th/11Yoshihiro Furukawa 55.0 560$ Mon Chouchou 1:55.5 <2 1/2> Nobu Osbourne <NK> Taisei Windy Ow. Tatsue Ishikawa 4,600,000 S 00000 Life50113M 30102 1 B 56.0 Genki Maruyama(8.2%,18−14−20−168,11th) Turf40112 I 20011 2 Trigger(JPN) Dirt10001L 00000 0Grass Wonder(0.59) 0Sunday Silence C3,b. -

Sake Spectator SAKE DATA BANK 2015 27
Sake Spectator SAKE DATA BANK 2015 27 Junmai Daiginjo 21 Grams Azumaichi BORN BORN Dassai 39 Dassai 50 Denemon Denshin Gasanryu Junmai Junmai Daiginjo Junmai Daiginjo Dreams Come True Gold Junmai Daiginjo Junmai Daiginjo Junmai Daiginjo Rin Gokugetsu Daiginjo Junmai Daiginjo Daiginjo Daiginjo Junmai Ginjo Junmai Ginjo Class Junmai Daiginjo Class Junmai Daiginjo Class Junmai Daiginjo Class Junmai Daiginjo Class Junmai Daiginjo Class Junmai Daiginjo Class Junmai Daiginjo Class Junmai Daiginjo Class Junmai Daiginjo Brewery Brewery Brewery Brewery Brewery Brewery Brewery Brewery Brewery Ginjo Abekan Shuzouten Gochouda Brewery Katoukichibee Shouten Katoukichibee Shouten Asahi Shuzo Asahi Shuzo Echigo Denemon Ippongi Kubohonten Shindo Sake Brewery Distributor Distributor Distributor Distributor Distributor Distributor Distributor Distributor Distributor Wismettac Asian Foods Banzai Beverage Corporation Mutual Trading Co., Inc. Mutual Trading Co., Inc. Mutual Trading Co., Inc. Mutual Trading Co., Inc. Wismettac Asian Foods JFC International Inc JFC International Inc Ginjo (562) 802-1900 (310) 634-9898 (213) 626-9458 (213) 626-9458 (213) 626-9458 (213) 626-9458 (562) 802-1900 (323) 721-6100 (323) 721-6100 Junmai/Tokubetsu Junmai www.abekan.com www.azumaichi.com www.lamtc.com www.lamtc.com www.lamtc.com www.lamtc.com denemon.com www.sakeexpert.com www.sakeexpert.com BORN BORN Daishichi Daishichi Hakushika Junmai/Tokubetsu Junmai Honjozo/Tokubetsu Honjozo Dassai 23 Gekkeikan Hakutsuru Hananomai Muroka Tokusen Houreki Minowamon Ginban Banshu 50 Sennenju -

Nord-Honshū (Tōhoku)
TM Nord-Honshū (Tōhoku) Tōhoku Vorwort Selbst Besuchern, die schon in Japan waren, ist die Region Tōhoku, die den nördlichen Teil der Hauptinsel Honshū umfasst, oft gänzlich unbekannt. Dabei findet man hier all das, was Japan so faszinierend macht, und zudem ohne Besuchermassen. Bis vor kurzem war der Norden Honshūs tatsächlich schwierig zu bereisen – die öffentlichen Verkehrsmittel waren nicht so gut ausgebaut wie im südlichen Teil von Honshū, und auch die englisch- sprachige Ausschilderung ließ noch zu wünschen übrig. Dies alles ist nun im Vorfeld der Olympischen Sommerspiele in Tokio 2020 in Angriff genommen worden. Tōhoku begeistert mit großartigen Landschaften wie den Heiligen Drei Bergen Dewa San- zan in Yamagata, der berühmten Bucht von Matsushima nahe der Stadt Sendai, oder mit der Mondlandschaft um den Vulkan Osorezan. Für kulturell Interessierte bieten die UNESCO Weltkulturerbe-Stätten von Hiraizumi einen Einblick in vergangene Feudalzei- ten. Zahlreiche Feste wie das berühmte Nebuta Matsuri in Aomori oder das Reiter-Festi- val Soma Nomaoi locken jedes Jahr zigtausende Besucher in die Region. FASZINIEREND — BUNT — JAPAN Kurz: es gibt hier Vieles zu entdecken in einer bisher wenig besuchten Region Japans. Das findet inzwischen auch Lonely Planet: für das Jahr 2020 steht Tōhoku an dritter Stelle WIR FLIEGEN SIE HIN! unter den vom Lonely Planet an Top 10 gesetzten Regionen weltweit. Mit diesem Booklet möchten wir Ihren Appetit wecken – schauen Sie selbst, was Tōhoku Ihnen alles zu bieten GOEntdecken Sie Japans viele Facetten und tauchen Sie hat! ein in unvergessliche Erlebnisse. Viel Freude beim Entdecken wünscht Ihnen das JNTO Frankfurt-Team! Erleben Sie Japan bereits bei uns an Bord — 4x täglich und nonstop mit ANA von Deutschland nach Tokio und darüber hinaus. -

Jprail-Timetable-Kamiki.Pdf
JPRail.com train timetable ThisJPRail.com timetable contains the following trains: JPRail.com 1. JR limited express trains in Hokkaido, Hokuriku, Chubu and Kansai area 2. Major local train lines in Hokkaido 3. Haneda airport access trains for the late arrival and the early departure 4. Kansai airport access trains for the late arrival and the early departure You may find the train timetables in the links below too. 1. JR East official timetable (Hokkaido, Tohoku, Akita, Yamagata, Joetsu and Hokuriku Shinkansens, Narita Express, Limited Express Azusa, Odoriko, Hitachi, Nikko, Inaho, and major joyful trains in eastern Japan) 2.JPRail.comJR Kyushu official timetable (Kyushu Shinkansen, JPRail.com Limited Express Yufuin no Mori, Sonic, Kamome, and major D&S trains) Timetable index (valid until mid March 2020) *The timetables may be changed without notice. The Limited Express Super Hokuto (Hakodate to Sapporo) 4 The Limited Express Super Hokuto (Sapporo to Hakodate) 5 JPRail.comThe Limited Express Suzuran (Muroran to Sapporo) JPRail.com6 The Limited Express Suzuran (Sapporo to Muroran) 6 The Limited Express Kamui and Lilac (Sapporo to Asahikawa) 7 The Limited Express Kamui and Lilac (Asahikawa to Sapporo) 8 The Limited Express Okhotsk and Taisetsu 9 The Limited Express Super Ozora and Super Tokachi (Sapporo to Obihiro and Kushiro) 10 The Limited Express Super Ozora and Super Tokachi (Obihiro and Kushiro to Sapporo) 11 JPRail.comThe Limited Express Soya and Sarobetsu JPRail.com12 Hakodate line local train between Otaru and Oshamambe (Oshamambe -

Japan Rail Pass Nas 02
01 02 03 01. Você pode adquirir o Japan Rail Pass nas 02. Tipos e preço da JAPAN RAIL PASS: Japan Rail Pass condições 1 e 2 , como está abaixo: Existem dois tipos de JAPAN RAIL PASS: Turistas estrangeiros visitando o Japão com visto a) Green: para vagões verdes - primeira classe. 1 b) Comum: classe turística. temporário (15 ou 90 dias). A JRP é oferecido pelas seis empresas que compõem o grupo Tem passe de 7 dias, 14 dias, ou de 21 dias. Japan Railways (JR Group) De acordo com as Leis da Imigração Japonesa, o status “Visi- Tipo Green Comum tante Temporário“ permite a permanência no Japão de 15 ou 90 dias. Ao entrar no Japão solicitando a permanência para turismo Duração Adulto Criança Adulto Criança 7dias ¥ ¥ ¥ ¥ JR九州 JR四国 JR西日本 JR東海 JR東日本 JR北海道 o funcionário irá carimbar seu passaporte como “TEMPORARY 38,880 19,440 29,110 14,550 (JR-Kyushu) (JR-Shikoku) (JR-Nishi-Nippon) (JR-Toukai) (JR-Higashi- (JR-Hokkaido) VISITOR”, como aparece abaixo. Somente as pessoas portando Nippon) 14dias ¥62,950 ¥31,470 ¥46,390 ¥23,190 o passaporte com este carimbo podem usar o PASSE DE TREM O JAPAN RAIL PASS oferece uma maneira incrivelmente JAPONÊS. 21 dias ¥81,870 ¥40,930 ¥59,350 ¥29,670 econômica de viajar sem limites por todo o Japão pela via férrea. Crianças de 6 à 11anos. * Valores válidos a partir de 01 / Abril / 2014. Esteja ciente, no entanto que se aplicam algumas restrições. O No Brasil os valores são convertidas pelo câmbio do dia, do iene ao passe não é válida para Trem bala (*)“Nozomi” e “Mizuho” que dollar, e do dollar serão convertidas em reais. -
Jr East Group Csr Report 2017
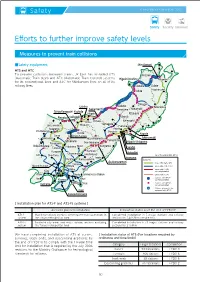
Safety JR EAST GROUP CSR REPORT 2017 Efforts to further improve safety levels Measures to prevent train collisions ■Safety equipment Shin-Aomori ATS and ATC To prevent collisions between trains, JR East has installed ATS Hirosaki (Automatic Train Stop) and ATC (Automatic Train Control) systems Higashi-Noshiro Aomori for its conventional lines and ATC for Shinkansen lines on all of its railway lines. Oiwake Ōdate Akita Hachinohe Ōmagari Uchino Sakata Morioka Sakamachi Amarume Hanamaki Echigo-Kawaguchi Yoshida Niigata Kitakami Naoetsu Kashiwazaki Shinjō Ichinoseki Niitsu Shibata Yamagata Jōetsumyōkō Miyauchi Furukawa Nagano Ayashi Kita-Matsumoto Shiroishi Koide Kogota Ōmae Echigo-Yuzawa Matsumoto Aizu-Wakamatsu Ishinomaki Fukushima Minakami Higashi-Shiogama Yokokawa Shin-Shirakawa Kobuchizawa Shibukawa Nikkō Kōriyama Takasaki Sendai Kuragano Kuroiso Utsunomiya Aobadōri Kōfu Oku-Tama Hōshakuji (as of the end of Mar. 2017) Iwaki Iwanuma Oyama Karasuyama 【Legend】 Asakanagamori : Lines with digital ATC Tomobe Musashi-Itsukaichi Ōmiya : Lines with ATC, ATS-P Abiko Mito : Lines with ATACS Kōzu currently installed Atami Narita Kashima-Soccer Stadium : Lines with ATS-Ps : Stations with ATS-P Itō Kisarazu Katori currently installed Narutō Chōshi : Stations with ATS-Ps Kurihama currently installed : Stations planned to be Ōami equipped with ATS-P Kazusa-Kameyama [ Installation plan for ATS-P and ATS-Ps systems ] Areas for planned installation Installation status as of the end of FY2017 ATS-P Mainly for railway sections with frequent train operations -

Annual Report2007
A Brief History of JR East 1987 April 1 • JR East established through division and privati- 1989 April 1 • Safety Research Laboratory and General zation of JNR Training Center established • Tokyo Regional Operations Headquarters, • Fares revised in connection with introduction of Tokyo Region Marketing Headquarters, Tohoku Japan’s national consumption tax Regional Headquarters, Niigata Branch, and May 20 • New-type ATS-P (Automatic Train Stop) devices Nagano Branch established introduced to enhance safety April 9 • Railway Safety Promotion Committee meeting October 23 • JR East InfoLine English-language information convened for the first time service began April 24 • American Potato new-style directly operated December 1 • ATS-SN devices introduced beer garden restaurant opened in Shimbashi Station 1990 March 7 • First Safety Seminar held May 20 • Casualty insurance agency business begun March 10 • Timetable revised May 25 • Catch phrase “From your neighborhood all the • Tokyo–Soga section of Keiyo Line opened way to the future” adopted March 25 • ATS-P use begun on Tokyo–Nakano section of June 7 • Green Counter customer feedback desk opened Chuo Rapid Line and Nakano–Chiba section of Chuo/Sobu Local Line July 1 • Domestic travel marketing business begun April 1 • Morioka Office and Akita Office upgraded to July 21 • Tokyo Ekikon station concert series begun branches October 1 • Huasa di Croma voluntary tip system rest room April 28 • Resort Limited Express Super View Odoriko facility opened at Shimbashi Station debuted October 15 • General -

Information on Suspension of Services Inside Shinkansen and Other Limited Express Trains on Conventional Lines
January 13th, 2021 East Japan Railway Co., Ltd. Information on suspension of services inside Shinkansen and other limited express trains on conventional lines We will suspend sales services inside Shinkansen cars and other limited express trains on conventional lines in order to prevent the spread of the novel coronavirus. Details on the suspension are as follows. We will notify customers when we resume the service. 1.Suspension period From January 16, 2021 (Sat) until the time being 2.Suspended services (1) Gran Class service in the Tohoku, Hokkaido, Joetsu, Hokuriku Shinkansen. ・ Gran Class sales will be canceled on all the trains starting from January 16, 2021 (Sat). ・ Beverage and snack services provided by attendants at Gran Class will be suspended on the following trains. 〇Tohoku and Hokkaido Shinkansen ・ Hayabusa(Section: Tokyo to Morioka, Shin-Aomori, and Shin-Hakodate-Hokuto) ・ Yamabiko(Section: Tokyo to Morioka) *There is no beverage and snack service on Hayabusa and Yamabiko in which the start and terminal station is Sendai. ○Hokuriku Shinkansen ・ Kagayaki(Section:Tokyo to Kanazawa) ・ Hakutaka(Section:Tokyo to Kanazawa) *To customers who already bought Gran Class tickets (January 16 to February 13) ・Gran Class (with beverage and snack service) Because we will suspend sales of the Gran Class, please change your ticket to Green Car ticket in advance. (The difference between Gran Class and Green Car will be reimbursed) ・Gran Class without beverage and snack service As the service content remains the same, only customers who have this ticket should use the reserved seat. (2)Sales inside Shinkansen, limited express trains on conventional lines, and Green Cars on regular trains. -

Origins of Traditional Cultivars of Primula Sieboldii Revealed by Nuclear Microsatellite and Chloroplast DNA Variations
Breeding Science 58: 347–354 (2008) Origins of traditional cultivars of Primula sieboldii revealed by nuclear microsatellite and chloroplast DNA variations Masanori Honjo1,2), Takashi Handa3), Yoshihiko Tsumura4), Izumi Washitani1) and Ryo Ohsawa*3) 1) Graduate School of Agricultural and Life Science, The University of Tokyo, 1-1-1 Yayoi, Bunkyo, Tokyo 113-8657, Japan 2) National Agriculture Research Center for Tohoku Region, 92 Shimokuriyagawa-Nabeyashiki, Morioka, Iwate 020-0123, Japan 3) Graduate School of Life and Environmental Sciences, University of Tsukuba, 1-1-1 Tennoudai, Tsukuba, Ibaraki 305-8572, Japan 4) Department of Forest Genetics, Forestry and Forest Products Research Institute, 1 Matsunosato, Tsukuba, Ibaraki 305-8687, Japan We examined the origins of 120 cultivars of Primula sieboldii, a popular Japanese pot plant with a cultivation history of more than 300 years. In an assignment test based on the microsatellite allelic composition of rep- resentative wild populations of P. sieboldii from the Hokkaido to Kyushu regions of Japan, most cultivars showed the highest likelihood of derivation from wild populations in the Arakawa River floodplain. Chloro- plast DNA haplotypes of cultivars also suggested that most cultivars have come from genets originating in wild populations from the same area, but, in addition, that several are descended from genets originating in other regions. The existence of three haplotypes that have not been found in current wild populations suggests that traditional cultivars may retain genetic diversity lost from wild populations. Key Words: assignment test, gene bank, genetic resource, haplotype, horticulture. Introduction 1985). These traditional cultivars of P. sieboldii are believed to have been established by finding genets with an uncom- Knowledge of the origins of cultivated plant species can be mon appearance in wild populations, and by subsequent in- useful in understanding the evolution of crop species traspecific crossing (Torii 1985). -

Company Pro Le
Company Prole global.kawasaki.com Cat.No.1A0296 Sep. '20 M Powering Table of Contents 3 Message from the President your 5 History 7 Kawasaki Hydrogen Road potential 9 Ship & Offshore Structure Company The Kawasaki Group creates new value by channeling its 11 Rolling Stock Company engineering prowess into various elds, including aerospace 13 Aerospace Systems Company systems, energy systems and plant engineering, precision 15 Energy System & Plant Engineering Company machinery and robots, and transportation, and also by pursuing 17 Motorcycle & Engine Company synergy that goes beyond the boundaries of these respective 19 Precision Machinery & Robot Company elds. Kawasaki strives to maintain harmony with the global 19 Precision Machinery Business Division environment as it works toward its vision of a better future. 21 Robot Business Division 23 Research & Development 25 Domestic Production and Sales Bases 27 Overseas Production Bases 29 Social Contribution 30 Information, Corporate Data Message from the President Kawasaki: A Corporate Group that Aptly Evaluates Social Needs and Swiftly Accommodates Changes The world is currently undergoing a major paradigm shift Since its incorporation in 1896, for more than 120 value for our customers. We therefore set “Trustworthy driven by an array of factors. These include an increased years, the Kawasaki Group has been developing Solutions for the Future” as our vision to be achieved by risk of environmental deterioration due to rapid industrial sophisticated technologies and generating knowledge 2030. This vision expresses our commitment to “making development and population expansion in emerging used for manufacturing products that encompass the land, available in a timely manner innovative solutions which economies, shrinking workforces as a result of “graying” sea, and air sectors.