Identification and Characterization of the Forkhead Box
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Rise and Fall of the Bovine Corpus Luteum
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Spring 5-6-2017 The Rise and Fall of the Bovine Corpus Luteum Heather Talbott University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Biochemistry Commons, Molecular Biology Commons, and the Obstetrics and Gynecology Commons Recommended Citation Talbott, Heather, "The Rise and Fall of the Bovine Corpus Luteum" (2017). Theses & Dissertations. 207. https://digitalcommons.unmc.edu/etd/207 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. THE RISE AND FALL OF THE BOVINE CORPUS LUTEUM by Heather Talbott A DISSERTATION Presented to the Faculty of the University of Nebraska Graduate College in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biochemistry and Molecular Biology Graduate Program Under the Supervision of Professor John S. Davis University of Nebraska Medical Center Omaha, Nebraska May, 2017 Supervisory Committee: Carol A. Casey, Ph.D. Andrea S. Cupp, Ph.D. Parmender P. Mehta, Ph.D. Justin L. Mott, Ph.D. i ACKNOWLEDGEMENTS This dissertation was supported by the Agriculture and Food Research Initiative from the USDA National Institute of Food and Agriculture (NIFA) Pre-doctoral award; University of Nebraska Medical Center Graduate Student Assistantship; University of Nebraska Medical Center Exceptional Incoming Graduate Student Award; the VA Nebraska-Western Iowa Health Care System Department of Veterans Affairs; and The Olson Center for Women’s Health, Department of Obstetrics and Gynecology, Nebraska Medical Center. -

The Functional Parasitic Worm Secretome: Mapping the Place of Onchocerca Volvulus Excretory Secretory Products
pathogens Review The Functional Parasitic Worm Secretome: Mapping the Place of Onchocerca volvulus Excretory Secretory Products Luc Vanhamme 1,*, Jacob Souopgui 1 , Stephen Ghogomu 2 and Ferdinand Ngale Njume 1,2 1 Department of Molecular Biology, Institute of Biology and Molecular Medicine, IBMM, Université Libre de Bruxelles, Rue des Professeurs Jeener et Brachet 12, 6041 Gosselies, Belgium; [email protected] (J.S.); [email protected] (F.N.N.) 2 Molecular and Cell Biology Laboratory, Biotechnology Unit, University of Buea, Buea P.O Box 63, Cameroon; [email protected] * Correspondence: [email protected] Received: 28 October 2020; Accepted: 18 November 2020; Published: 23 November 2020 Abstract: Nematodes constitute a very successful phylum, especially in terms of parasitism. Inside their mammalian hosts, parasitic nematodes mainly dwell in the digestive tract (geohelminths) or in the vascular system (filariae). One of their main characteristics is their long sojourn inside the body where they are accessible to the immune system. Several strategies are used by parasites in order to counteract the immune attacks. One of them is the expression of molecules interfering with the function of the immune system. Excretory-secretory products (ESPs) pertain to this category. This is, however, not their only biological function, as they seem also involved in other mechanisms such as pathogenicity or parasitic cycle (molting, for example). Wewill mainly focus on filariae ESPs with an emphasis on data available regarding Onchocerca volvulus, but we will also refer to a few relevant/illustrative examples related to other worm categories when necessary (geohelminth nematodes, trematodes or cestodes). -

Targeting EZH2 Reactivates a Breast Cancer Subtype-Specific Anti-Metastatic Transcriptional Program
ARTICLE DOI: 10.1038/s41467-018-04864-8 OPEN Targeting EZH2 reactivates a breast cancer subtype-specific anti-metastatic transcriptional program Alison Hirukawa1,2, Harvey W. Smith1, Dongmei Zuo1, Catherine R. Dufour1, Paul Savage 1, Nicholas Bertos 1, Radia M. Johnson1, Tung Bui1,2, Guillaume Bourque3,4, Mark Basik5,6, Vincent Giguère 1,2,6, Morag Park1,2,6 & William J. Muller1,2,6 1234567890():,; Emerging evidence has illustrated the importance of epigenomic reprogramming in cancer, with altered post-translational modifications of histones contributing to pathogenesis. However, the contributions of histone modifiers to breast cancer progression are unclear, and how these processes vary between molecular subtypes has yet to be adequately addressed. Here we report that genetic or pharmacological targeting of the epigenetic modifier Ezh2 dramatically hinders metastatic behaviour in both a mouse model of breast cancer and patient-derived xenografts reflective of the Luminal B subtype. We further define a subtype- specific molecular mechanism whereby EZH2 maintains H3K27me3-mediated repression of the FOXC1 gene, thereby inactivating a FOXC1-driven, anti-invasive transcriptional program. We demonstrate that higher FOXC1 is predictive of favourable outcome specifically in Luminal B breast cancer patients and establish the use of EZH2 methyltransferase inhibitors as a viable strategy to block metastasis in Luminal B breast cancer, where options for targeted therapy are limited. 1 Goodman Cancer Research Centre, McGill University, Montréal, QC H3A 1A3, Canada. 2 Department of Biochemistry, McGill University, Montréal, QC H3G 1Y6, Canada. 3 McGill University, Génome Québec Innovation Centre, Montréal, QC H3A 0G1, Canada. 4 Department of Human Genetics, McGill University, Montréal, QC H3A 1A3, Canada. -
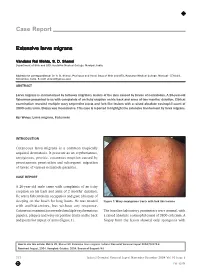
Extensive Larva Migrans
Case Report Extensive larva migrans Vandana Rai Mehta, S. D. Shenoi Department of Skin and STD, Kasturba Medical College, Manipal, India. Address for correspondence: Dr. S. D. Shenoi, Professor and Head, Dept of Skin and STD, Kasturba Medical College, Manipal - 576104, Karnataka, India. E-mail: [email protected] ABSTRACT Larva migrans is characterized by tortuous migratory lesions of the skin caused by larvae of nematodes. A 26-year-old fisherman presented to us with complaints of an itchy eruption on his back and arms of two months’ duration. Clinical examination revealed multiple wavy serpentine tracts and fork like lesions with a raised absolute eosinophil count of 3800 cells/cmm. Biopsy was inconclusive. This case is reported to highlight the extensive involvement by larva migrans. KEY WORDS: Larva migrans, Fisherman INTRODUCTION Cutaneous larva migrans is a common tropically acquired dermatosis. It presents as an erythematous, serpiginous, pruritic, cutaneous eruption caused by percutaneous penetration and subsequent migration of larvae of various nematode parasites. CASE REPORT A 26-year-old male came with complaints of an itchy eruption on his back and arms of 2 months’ duration. He was a fisherman by occupation and gave a history of sleeping on the beach for long hours. He was treated Figure 1: Wavy serpiginous tracts with fork like lesions with antihistamines, but without any response. Cutaneous examination revealed multiple erythematous The baseline laboratory parameters were normal, with papules, plaques and wavy serpentine tracts on the back a raised absolute eosinophil count of 3800 cell/cmm. A and posterior aspect of arms (Figure 1). biopsy from the lesion showed only spongiosis with How to cite this article: Mehta VR, Shenoi SD. -

Insecurities and Dogs: an Obstacle to the Eradication of Dracunculiasis
dicine & Me S l u a r ic g e p r o y r T ISSN: 2329-9088 Tropical Medicine & Surgery Review Article Insecurities and Dogs: An Obstacle to the Eradication of Dracunculiasis Aja Kalu1*,Nwufo Amanda2 Department of Care of the Elderly, Barking, Havering and Redbridge University Hospital, NHS Trust, Romford, Essex, UK; 2 Department of Public Health, University of Chester, Chester, UK. ABSTRACT Dracunculiasis is a parasitic worm infection also known as Guinea Worm Disease (GWD). It is caused by a nematode called Dracunculiasis Medinensis. It belongs to a group of communicable disease named Neglected Tropic Disease (NTD). Dracunculiasis is caused by drinking water contaminated with the vector copepods (water fleas). Although the disease is not fatal, the sores caused by the emerging worm in the lower limb can become secondarily infected and complications such as sepsis, tetanus can ensue. Also, the sores can cause abscess and cellulitis, leaving the individual incapacitated for weeks which extends beyond the emergence of the worm. Over the last three decades, the prevalence of Guinea worm disease has reduced drastically through cost effective intervention provided by The Cater Center, WHO, UNICEF with the disease targeted for eradication. Some African countries like Nigeria, Ghana, South Africa, and Kenya being the most recent, have eliminated the disease. Guinea worm is still present in Chad, Cameroon, Mali, Ethiopia where political instability, social inequalities and infection of dogs by the worm pose an increasing threat and obstacle to the elimination of the disease. Dracunculiasis represents a disease that can be eradicated without a drug or vaccine but with a cost-effective intervention that involves community efforts. -

The Forkhead-Box Family of Transcription Factors: Key Molecular Players in Colorectal Cancer Pathogenesis Paul Laissue
Laissue Molecular Cancer (2019) 18:5 https://doi.org/10.1186/s12943-019-0938-x REVIEW Open Access The forkhead-box family of transcription factors: key molecular players in colorectal cancer pathogenesis Paul Laissue Abstract Colorectal cancer (CRC) is the third most commonly occurring cancer worldwide and the fourth most frequent cause of death having an oncological origin. It has been found that transcription factors (TF) dysregulation, leading to the significant expression modifications of genes, is a widely distributed phenomenon regarding human malignant neoplasias. These changes are key determinants regarding tumour’s behaviour as they contribute to cell differentiation/proliferation, migration and metastasis, as well as resistance to chemotherapeutic agents. The forkhead box (FOX) transcription factor family consists of an evolutionarily conserved group of transcriptional regulators engaged in numerous functions during development and adult life. Their dysfunction has been associated with human diseases. Several FOX gene subgroup transcriptional disturbances, affecting numerous complex molecular cascades, have been linked to a wide range of cancer types highlighting their potential usefulness as molecular biomarkers. At least 14 FOX subgroups have been related to CRC pathogenesis, thereby underlining their role for diagnosis, prognosis and treatment purposes. This manuscript aims to provide, for the first time, a comprehensive review of FOX genes’ roles during CRC pathogenesis. The molecular and functional characteristics of most relevant FOX molecules (FOXO, FOXM1, FOXP3) have been described within the context of CRC biology, including their usefulness regarding diagnosis and prognosis. Potential CRC therapeutics (including genome-editing approaches) involving FOX regulation have also been included. Taken together, the information provided here should enable a better understanding of FOX genes’ function in CRC pathogenesis for basic science researchers and clinicians. -

Public Health Significance of Intestinal Parasitic Infections*
Articles in the Update series Les articles de la rubrique give a concise, authoritative, Le pointfournissent un bilan and up-to-date survey of concis et fiable de la situa- the present position in the tion actuelle dans les do- Update selectedfields, coveringmany maines consideres, couvrant different aspects of the de nombreux aspects des biomedical sciences and sciences biomedicales et de la , po n t , , public health. Most of santepublique. Laplupartde the articles are written by ces articles auront donc ete acknowledged experts on the redigeis par les specialistes subject. les plus autorises. Bulletin of the World Health Organization, 65 (5): 575-588 (1987) © World Health Organization 1987 Public health significance of intestinal parasitic infections* WHO EXPERT COMMITTEE' Intestinal parasitic infections are distributed virtually throughout the world, with high prevalence rates in many regions. Amoebiasis, ascariasis, hookworm infection and trichuriasis are among the ten most common infections in the world. Other parasitic infections such as abdominal angiostrongyliasis, intestinal capil- lariasis, and strongyloidiasis are of local or regional public health concern. The prevention and control of these infections are now more feasible than ever before owing to the discovery of safe and efficacious drugs, the improvement and sim- plification of some diagnostic procedures, and advances in parasite population biology. METHODS OF ASSESSMENT The amount of harm caused by intestinal parasitic infections to the health and welfare of individuals and communities depends on: (a) the parasite species; (b) the intensity and course of the infection; (c) the nature of the interactions between the parasite species and concurrent infections; (d) the nutritional and immunological status of the population; and (e) numerous socioeconomic factors. -

Research Progress on the Forkhead Box C1
www.impactjournals.com/oncotarget/ Oncotarget, 2018, Vol. 9, (No. 15), pp: 12471-12478 Review Research progress on the forkhead box C1 Jinhua Wang1,2, Wan Li2, Xiangjin Zheng2, Xiaocong Pang2 and Guanhua Du1,2 1The State Key Laboratory of Bioactive Substance and Function of Natural Medicines, Beijing 100050, China 2Key Laboratory of Drug Target Research and Drug Screen, Institute of Materia Medica, Chinese Academy of Medical Science and Peking Union Medical College, Beijing 100050, China Correspondence to: Jinhua Wang, email: [email protected] Guanhua Du, email: [email protected] Keywords: FOX family; FOXC1; cancer; drug resistance; stem cell Received: July 29, 2017 Accepted: November 01, 2017 Published: November 20, 2017 Copyright: Wang et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT FOXC1 is a vital member of FOX families which play important roles in biological processes including proliferation, differentiation, apoptosis, migration, invasion, metabolism, and longevity. Here we are focusing on roles of FOXC1 and their mechanisms in cancers. FOXC1 promoted progress of many cancers, such as breast cancer (especially basal-like breast cancer), hepatocellular carcinoma, gastric cancer and so on. FOXC1 was also found to be associated with drug resistance of cancers. FOXC1 promoted metastasis of cancers by increasing expression of MMP7, NEDD9 and Snail. Proliferation and invasion of cancers were increased by FOXC1 by mediating NF-κB, MST1R and KLF4 expression. FOXC1 was associated with development by regulating expression of FGF19 and MSX1. -

Risk of Soil-Transmitted Helminthiasis Among Agrarian Communities of Kogi
bioRxiv preprint doi: https://doi.org/10.1101/663237; this version posted June 7, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 1 Long title: 2 Risk of soil-transmitted helminthiasis among agrarian communities of Kogi 3 State, Nigeria: Evaluated in the context of The Soil-Transmitted 4 Helminthiasis Advisory Committee recommendation 2016 5 Short title: 6 Soil-transmitted helminthiasis in Kogi State 7 8 Joy T. Anunobi1, Ikem C. Okoye2*, Ifeanyi Oscar N. Aguzie2*, Yvonne E. Ndukwe2 and 9 Onyekachi J. Okpasuo2 10 1 Science Laboratory Technology Department, Federal Polytechnic, Idah, Kogi State, Nigeria. 11 2Parasitology and Public Health Unit, Department of Zoology and Environmental Biology, 12 University of Nigeria, Nsukka, Enugu State, Nigeria. 13 *Corresponding authors: 14 E-mail: [email protected] (IONA) 15 E-mail: [email protected] (ICO) 16 Authors’ contributions 17 Conceptualization and Methodology: Joy T. Anunobi and Ikem C. Okoye 18 Investigation: Joy T. Anunobi, Yvonne E. Ndukwe and Onyekachi J. Okpasuo 19 Data curation, formal analysis & software: Ifeanyi Oscar N. Aguzie 20 Writing of first draft: Joy T. Anunobi and Ifeanyi Oscar N. Aguzie 21 Supervision: Ikem C. Okoye 22 Final draft: All the authors approved the final draft. 23 Funding 24 The study did not receive external funding 1 bioRxiv preprint doi: https://doi.org/10.1101/663237; this version posted June 7, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

In Vitro and in Vivo Trematode Models for Chemotherapeutic Studies
589 In vitro and in vivo trematode models for chemotherapeutic studies J. KEISER* Department of Medical Parasitology and Infection Biology, Swiss Tropical Institute, CH-4002 Basel, Switzerland (Received 27 June 2009; revised 7 August 2009 and 26 October 2009; accepted 27 October 2009; first published online 7 December 2009) SUMMARY Schistosomiasis and food-borne trematodiases are chronic parasitic diseases affecting millions of people mostly in the developing world. Additional drugs should be developed as only few drugs are available for treatment and drug resistance might emerge. In vitro and in vivo whole parasite screens represent essential components of the trematodicidal drug discovery cascade. This review describes the current state-of-the-art of in vitro and in vivo screening systems of the blood fluke Schistosoma mansoni, the liver fluke Fasciola hepatica and the intestinal fluke Echinostoma caproni. Examples of in vitro and in vivo evaluation of compounds for activity are presented. To boost the discovery pipeline for these diseases there is a need to develop validated, robust high-throughput in vitro systems with simple readouts. Key words: Schistosoma mansoni, Fasciola hepatica, Echinostoma caproni, in vitro, in vivo, drug discovery, chemotherapy. INTRODUCTION by chemotherapy. However, only two drugs are currently available: triclabendazole against fascio- Thus far approximately 6000 species in the sub-class liasis and praziquantel against the other food-borne Digenea, phylum Platyhelminthes have been de- trematode infections and schistosomiasis (Keiser and scribed in the literature. Among them, only a dozen Utzinger, 2004; Keiser et al. 2005). Hence, there is a or so species parasitize humans. These include need for discovery and development of new drugs, the blood flukes (five species of Schistosoma), liver particularly in view of growing concern about re- flukes (Clonorchis sinensis, Fasciola gigantica, Fasciola sistance developing to existing drugs. -

Whole Exome Sequencing in Families at High Risk for Hodgkin Lymphoma: Identification of a Predisposing Mutation in the KDR Gene
Hodgkin Lymphoma SUPPLEMENTARY APPENDIX Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene Melissa Rotunno, 1 Mary L. McMaster, 1 Joseph Boland, 2 Sara Bass, 2 Xijun Zhang, 2 Laurie Burdett, 2 Belynda Hicks, 2 Sarangan Ravichandran, 3 Brian T. Luke, 3 Meredith Yeager, 2 Laura Fontaine, 4 Paula L. Hyland, 1 Alisa M. Goldstein, 1 NCI DCEG Cancer Sequencing Working Group, NCI DCEG Cancer Genomics Research Laboratory, Stephen J. Chanock, 5 Neil E. Caporaso, 1 Margaret A. Tucker, 6 and Lynn R. Goldin 1 1Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 2Cancer Genomics Research Laboratory, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 3Ad - vanced Biomedical Computing Center, Leidos Biomedical Research Inc.; Frederick National Laboratory for Cancer Research, Frederick, MD; 4Westat, Inc., Rockville MD; 5Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; and 6Human Genetics Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD, USA ©2016 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol.2015.135475 Received: August 19, 2015. Accepted: January 7, 2016. Pre-published: June 13, 2016. Correspondence: [email protected] Supplemental Author Information: NCI DCEG Cancer Sequencing Working Group: Mark H. Greene, Allan Hildesheim, Nan Hu, Maria Theresa Landi, Jennifer Loud, Phuong Mai, Lisa Mirabello, Lindsay Morton, Dilys Parry, Anand Pathak, Douglas R. Stewart, Philip R. Taylor, Geoffrey S. Tobias, Xiaohong R. Yang, Guoqin Yu NCI DCEG Cancer Genomics Research Laboratory: Salma Chowdhury, Michael Cullen, Casey Dagnall, Herbert Higson, Amy A.