A Two-Step Graph Convolutional Decoder for Molecule Generation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

On the Formation of Higher Carbon Oxides in Extreme Environments
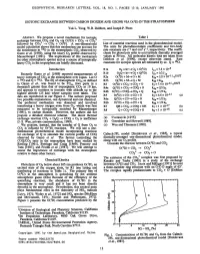
Chemical Physics Letters 465 (2008) 1–9 Contents lists available at ScienceDirect Chemical Physics Letters journal homepage: www.elsevier.com/locate/cplett FRONTIERS ARTICLE On the formation of higher carbon oxides in extreme environments Ralf I. Kaiser a,*, Alexander M. Mebel b a Department of Chemistry, University of Hawaii at Manoa, 2545 The Mall (Bil 301A), Honolulu, HI 96822, USA b Department of Chemistry and Biochemistry, Florida International University, Miami, FL 33199, USA article info abstract Article history: Due to the importance of higher carbon oxides of the general formula COx (x > 2) in atmospheric chem- Received 15 May 2008 istry, isotopic enrichment processes, low-temperature ices in the interstellar medium and in the outer In final form 22 July 2008 solar system, as well as potential implications to high-energy materials, an overview on higher carbon Available online 29 July 2008 oxides COx (x = 3–6) is presented. This article reviews recent developments on these transient species. Future challenges and directions of this research field are highlighted. Ó 2008 Elsevier B.V. All rights reserved. 1. Introduction tion has no activation energy, proceeds with almost unit efficiency, and most likely involves a reaction intermediate. However, neither In recent years, the interest in carbon oxides of higher complex- reaction products (kinetics studies) nor the nature of the interme- 1 + ityP than carbon monoxide (CO; X R ) and carbon dioxide (CO2; diate (kinetics and dynamicsP studies) were determined. On the 1 þ 1 + 3 À X g ) of the generic formula COx (x > 2) has been fueled by com- other hand, a CO(X R )+O2(X g ) exit channel was found to have plex reaction mechanisms of carbon oxides with atomic oxygen in an activation energy between 15 and 28 kJ molÀ1 in the range of the atmospheres of Mars [1–4] and Venus [5]. -
Isotopic Exchange Between Carbon Dioxide and Ozone Via O1D in The
GEOPHYSICAL RESEARCH LETTERS, VOL. 18, NO. 1, PAGES 13-16, JANUARY 1991 ISOTOPICEXCHANGE BETWEEN CARBONDIOXIDE AND OZONEVIA O(!D) IN THE STRATOSPHERE Yuk L. Yung, W.B. DoMore,and Joseph P. Pinto Abstract. We proposea novelmechanism for isotopic Table 1 exchangebetween CO 2 and0 3 vi•aO(ID) + CO2 -->CO_•* followedby CO3' --> CO2 + O(aP). A one-•ensior•al List of essentialreactions used in the photochemicalmodel. model calculation shows that this mechanism can account for The units for photodissociationcoefficients and two-body theenrichment in 180 in thestratospheric CO2 observed by rateconstants are s -1 andcm 3 s-l, respectively.The coeffi- Gatnoeta!. [1989],using the heavy 0 3 profile-observed by cientsfor photolysisrefer to nfid-latitudedimally averaged Mauersberger[1981]. The implicationsof this mechanism values at 30 kan. All molecular kinetic data are taken frmn for otherstratospheric species and as a sourceof isotopically DoMore etal. [1990], except otherwise stated. Rate heavyCO 2 in thetroposphere are briefly discussed. constantsforisotopic species are estimated byus. Q = 180. h•troduction Rla O3+ bY-->O2 + O(1D) Jla=7.4x10 -5 RecentlyGamo et at. [1989] reportedmeasurements of Rib O2Q+ hv-->O2 + Q(!D) Jlb=1/2 J la heavyisotopes of CO2 in the stratosphereover Japan. Let O R2a O(ID)+ M--->O+ M k2a= 2.0x 10-11 e ll0/T = 160 andQ = 180.The fiQ of stratosphericCO2, as defined R2b Q(1D) + M --->Q + M k2b= k2a in Gatno et at., was found to be 2O/oo(two parts in a R3 O(ID)+ CO2---->CO2 + O k3 = 7.4x 10-11 e 120/T thousand)greater than that of troposphericCO 2 at 19 km, R4a Q(1D)+ CO2-->CO Q + O k4a= 2/3k 3 and appearsto continueto increasewith altitudeup to the R4b O(ID)+ COQ-->CO2 + Q k4b= 1/3k 3 highestaltitude (25 km) where sampleswere taken. -

Assessing the Chemistry and Bioavailability of Dissolved Organic Matter from Glaciers and Rock Glaciers
RESEARCH ARTICLE Assessing the Chemistry and Bioavailability of Dissolved 10.1029/2018JG004874 Organic Matter From Glaciers and Rock Glaciers Special Section: Timothy Fegel1,2 , Claudia M. Boot1,3 , Corey D. Broeckling4, Jill S. Baron1,5 , Biogeochemistry of Natural 1,6 Organic Matter and Ed K. Hall 1Natural Resource Ecology Laboratory, Colorado State University, Fort Collins, CO, USA, 2Rocky Mountain Research 3 Key Points: Station, U.S. Forest Service, Fort Collins, CO, USA, Department of Chemistry, Colorado State University, Fort Collins, • Both glaciers and rock glaciers CO, USA, 4Proteomics and Metabolomics Facility, Colorado State University, Fort Collins, CO, USA, 5U.S. Geological supply highly bioavailable sources of Survey, Reston, VA, USA, 6Department of Ecosystem Science and Sustainability, Colorado State University, Fort Collins, organic matter to alpine headwaters CO, USA in Colorado • Bioavailability of organic matter released from glaciers is greater than that of rock glaciers in the Rocky Abstract As glaciers thaw in response to warming, they release dissolved organic matter (DOM) to Mountains alpine lakes and streams. The United States contains an abundance of both alpine glaciers and rock • ‐ The use of GC MS for ecosystem glaciers. Differences in DOM composition and bioavailability between glacier types, like rock and ice metabolomics represents a novel approach for examining complex glaciers, remain undefined. To assess differences in glacier and rock glacier DOM we evaluated organic matter pools bioavailability and molecular composition of DOM from four alpine catchments each with a glacier and a rock glacier at their headwaters. We assessed bioavailability of DOM by incubating each DOM source Supporting Information: with a common microbial community and evaluated chemical characteristics of DOM before and after • Supporting Information S1 • Data Set S1 incubation using untargeted gas chromatography–mass spectrometry‐based metabolomics. -

Potential Ivvs.Gelbasedre
US010644304B2 ( 12 ) United States Patent ( 10 ) Patent No.: US 10,644,304 B2 Ein - Eli et al. (45 ) Date of Patent : May 5 , 2020 (54 ) METHOD FOR PASSIVE METAL (58 ) Field of Classification Search ACTIVATION AND USES THEREOF ??? C25D 5/54 ; C25D 3/665 ; C25D 5/34 ; ( 71) Applicant: Technion Research & Development HO1M 4/134 ; HOTM 4/366 ; HO1M 4/628 ; Foundation Limited , Haifa ( IL ) (Continued ) ( 72 ) Inventors : Yair Ein - Eli , Haifa ( IL ) ; Danny (56 ) References Cited Gelman , Haifa ( IL ) ; Boris Shvartsev , Haifa ( IL ) ; Alexander Kraytsberg , U.S. PATENT DOCUMENTS Yokneam ( IL ) 3,635,765 A 1/1972 Greenberg 3,650,834 A 3/1972 Buzzelli ( 73 ) Assignee : Technion Research & Development Foundation Limited , Haifa ( IL ) (Continued ) ( * ) Notice : Subject to any disclaimer , the term of this FOREIGN PATENT DOCUMENTS patent is extended or adjusted under 35 CN 1408031 4/2003 U.S.C. 154 ( b ) by 56 days . EP 1983078 10/2008 (21 ) Appl. No .: 15 /300,359 ( Continued ) ( 22 ) PCT Filed : Mar. 31 , 2015 OTHER PUBLICATIONS (86 ) PCT No .: PCT/ IL2015 /050350 Hagiwara et al. in ( Acidic 1 - ethyl - 3 -methylimidazoliuum fluoride: a new room temperature ionic liquid in Journal of Fluorine Chem $ 371 (c ) ( 1 ), istry vol . 99 ( 1999 ) p . 1-3 ; ( Year: 1999 ). * (2 ) Date : Sep. 29 , 2016 (Continued ) (87 ) PCT Pub . No .: WO2015 / 151099 Primary Examiner — Jonathan G Jelsma PCT Pub . Date : Oct. 8 , 2015 Assistant Examiner Omar M Kekia (65 ) Prior Publication Data (57 ) ABSTRACT US 2017/0179464 A1 Jun . 22 , 2017 Disclosed is a method for activating a surface of metals , Related U.S. Application Data such as self- passivated metals , and of metal -oxide dissolu tion , effected using a fluoroanion -containing composition . -

Chemical Names and CAS Numbers Final
Chemical Abstract Chemical Formula Chemical Name Service (CAS) Number C3H8O 1‐propanol C4H7BrO2 2‐bromobutyric acid 80‐58‐0 GeH3COOH 2‐germaacetic acid C4H10 2‐methylpropane 75‐28‐5 C3H8O 2‐propanol 67‐63‐0 C6H10O3 4‐acetylbutyric acid 448671 C4H7BrO2 4‐bromobutyric acid 2623‐87‐2 CH3CHO acetaldehyde CH3CONH2 acetamide C8H9NO2 acetaminophen 103‐90‐2 − C2H3O2 acetate ion − CH3COO acetate ion C2H4O2 acetic acid 64‐19‐7 CH3COOH acetic acid (CH3)2CO acetone CH3COCl acetyl chloride C2H2 acetylene 74‐86‐2 HCCH acetylene C9H8O4 acetylsalicylic acid 50‐78‐2 H2C(CH)CN acrylonitrile C3H7NO2 Ala C3H7NO2 alanine 56‐41‐7 NaAlSi3O3 albite AlSb aluminium antimonide 25152‐52‐7 AlAs aluminium arsenide 22831‐42‐1 AlBO2 aluminium borate 61279‐70‐7 AlBO aluminium boron oxide 12041‐48‐4 AlBr3 aluminium bromide 7727‐15‐3 AlBr3•6H2O aluminium bromide hexahydrate 2149397 AlCl4Cs aluminium caesium tetrachloride 17992‐03‐9 AlCl3 aluminium chloride (anhydrous) 7446‐70‐0 AlCl3•6H2O aluminium chloride hexahydrate 7784‐13‐6 AlClO aluminium chloride oxide 13596‐11‐7 AlB2 aluminium diboride 12041‐50‐8 AlF2 aluminium difluoride 13569‐23‐8 AlF2O aluminium difluoride oxide 38344‐66‐0 AlB12 aluminium dodecaboride 12041‐54‐2 Al2F6 aluminium fluoride 17949‐86‐9 AlF3 aluminium fluoride 7784‐18‐1 Al(CHO2)3 aluminium formate 7360‐53‐4 1 of 75 Chemical Abstract Chemical Formula Chemical Name Service (CAS) Number Al(OH)3 aluminium hydroxide 21645‐51‐2 Al2I6 aluminium iodide 18898‐35‐6 AlI3 aluminium iodide 7784‐23‐8 AlBr aluminium monobromide 22359‐97‐3 AlCl aluminium monochloride -

Carbon Dioxide - Wikipedia
5/20/2020 Carbon dioxide - Wikipedia Carbon dioxide Carbon dioxide (chemical formula CO2) is a colorless gas with Carbon dioxide a density about 60% higher than that of dry air. Carbon dioxide consists of a carbon atom covalently double bonded to two oxygen atoms. It occurs naturally in Earth's atmosphere as a trace gas. The current concentration is about 0.04% (412 ppm) by volume, having risen from pre-industrial levels of 280 ppm.[8] Natural sources include volcanoes, hot springs and geysers, and it is freed from carbonate rocks by dissolution in water and acids. Because carbon dioxide is soluble in water, it occurs naturally in groundwater, rivers and lakes, ice caps, glaciers and seawater. It is present in deposits of petroleum and natural gas. Carbon dioxide is odorless at normally encountered concentrations, but at high concentrations, it has a sharp and acidic odor.[1] At such Names concentrations it generates the taste of soda water in the Other names [9] mouth. Carbonic acid gas As the source of available carbon in the carbon cycle, atmospheric Carbonic anhydride carbon dioxide is the primary carbon source for life on Earth and Carbonic oxide its concentration in Earth's pre-industrial atmosphere since late Carbon oxide in the Precambrian has been regulated by photosynthetic organisms and geological phenomena. Plants, algae and Carbon(IV) oxide cyanobacteria use light energy to photosynthesize carbohydrate Dry ice (solid phase) from carbon dioxide and water, with oxygen produced as a waste Identifiers product.[10] CAS Number 124-38-9 (http://ww w.commonchemistr CO2 is produced by all aerobic organisms when they metabolize carbohydrates and lipids to produce energy by respiration.[11] It is y.org/ChemicalDeta returned to water via the gills of fish and to the air via the lungs of il.aspx?ref=124-38- air-breathing land animals, including humans. -

Standard Heats of Formation of Selected Compounds
Standard heats of formation of selected compounds f f se se H H a a o o h h Compound P Formula (kJ/mol) Compound P Formula (kJ/mol) Aluminium chloride Cesium ion + s AlCl3 705.63 g Cs 457.96 Aluminium oxide Cesium chloride s Al2O3 1669.8 s CsCl 443.04 Aluminium hydroxide Benzene s Al(OH)3 1277 l C6H6 48.95 Aluminium chloride Benzoic acid s Al2(SO4)3 3440 s C7H6O2 385.2 Barium chloride Glucose s BaCl2 858.6 s C6H12O6 1271 Barium carbonate Carbon (diamond) s BaCO3 1213 s C 1.90 Barium hydroxide Carbon gas s Ba(OH)2 944.7 g C 716.67 Barium oxide Carbon dioxide s BaO 548.1 g CO2 393.509 Barium sulfate Carbon disulfide liquid s BaSO4 1473.2 l CS2 89.41 Beryllium hydroxide Carbon disulfide gas s Be(OH)2 902.9999 g CS2 116.70 Beryllium oxide s BeO 609.4 Carbon monoxide g CO 110.525 Boron trichloride Carbon tetrachloride liquid s Bcl3 402.96 l CCl4 135.4 Bromine ion l Br 121 Carbon tetrachloride gas g CCl4 95.98 Bromine gas (monatomic) Phosgene g Br 111.88 g COCl2 218.8 Bromine gas (diatomic) Ethane g Br2 30.91 g C2H6 83.85 Bromine trifluoride Ethanol liquid g BrF3 255.60 l C2H5OH 277.0 Hydrobromic acid Ethanol gas g HBr 36.29 g C2H5OH 235.3 Cadmium oxide Ethene s CdO 258 g C2H4 52.30 Cadmium hydroxide Vinyl chloride s Cd(OH)2 561 s C2H3Cl 94.12 Cadmium sulfide Ethyne (acetylene) s CdS 162 g C2H2 226.73 Cadmium sulfate Methane s CdSO4 935 g CH4 74.87 Calcium gas Methanol liquid g Ca 178.2 l CH3OH 238.4 Calcium ion 2+ Methanol gas g Ca 1925.9 g CH3OH 201.0 Calcium carbide Methyl linoleate (biodiesel) s CaC2 59.8 g C19H34O2 356.3 Calcium carbonate (Calcite) -

Answer: 2 푦 푒 푥2 푥푒 Bonus
ROUND 13 TOSS-UP 1) MATH Short Answer Let = [e to the power of x squared]. Find dy/dx. 2 푥 ANSWER: 2 푦 푒 푥2 푥푒 BONUS 1) MATH Short Answer How many distinct permutations can be made from the letters in the word INFINITY? ANSWER: 3360 TOSS-UP 2) CHEMISTRY Multiple Choice What is the local VSEPR geometry of a fully substituted carbonyl carbon? W) Tetrahedral X) Trigonal pyramidal Y) Trigonal planar Z) T-shaped ANSWER: Y) TRIGONAL PLANAR BONUS 2) CHEMISTRY Multiple Choice Which of the following analytical tools can provide information about the crystalline structure of unknown materials? W) X-ray diffraction X) X-ray photoelectron spectroscopy Y) High pressure liquid chromatography Z) Dynamic light scattering ANSWER: W) X-RAY DIFFRACTION High School Round 13 A Page 1 TOSS-UP 3) PHYSICS Short Answer A ball is dropped from rest at a height of 60 meters above the ground. If the potential energy is zero at the ground, then at what height in meters is the potential energy 60% of its initial potential energy? ANSWER: 36 BONUS 3) PHYSICS Short Answer What is the minimum current in amperes a circuit breaker fuse must be able to handle in order to run a 1450 watt refrigerator hooked up to a 110 volt kitchen circuit, assuming fuses are only available in whole number increments? ANSWER: 14 TOSS-UP 4) EARTH AND SPACE Multiple Choice During an El Niño event, the thermocline [THUR-muh- klyn] in the eastern equatorial Pacific is which of the following, compared to normal? W) Thicker X) Thinner Y) The same Z) Either thicker or thinner depending on the salinity -
A Summary of Mission-Critical Cryogenic Laboratory Spectral Measurements for Determination of Icy Satellite Surface Composition from Orbital Spacecraft Observations
Science of Solar System Ices (2008) 9028.pdf A SUMMARY OF MISSION-CRITICAL CRYOGENIC LABORATORY SPECTRAL MEASUREMENTS FOR DETERMINATION OF ICY SATELLITE SURFACE COMPOSITION FROM ORBITAL SPACECRAFT OBSERVATIONS. J. B. Dalton, III1, 1Planetary Ices Group, Jet Propulsion Laboratory, MS 183- 301, 4800 Oak Grove Drive, Pasadena, CA 91109-8001. Introduction: The bulk of our knowledge regard- The overtones and combinations which make up most ing icy satellite surface composition is derived from of the VNIR spectral signatures are far weaker than the visible to near-infrared (VNIR) reflectance spectros- MIR fundamentals. However, reflected sunlight from copy, much of it from spacecraft observations. Spectra cold icy bodies in the outer solar system exhibits insuf- of planetary surfaces can be modeled either as linear ficient spectral contrast and inadequate signal to enable (areal) mixtures, or as nonlinear (intimate) mixtures, to the identification of surface materials in the MIR so yield estimates of relative abundance of surface com- remote-sensing instrumentation for icy bodies concen- pounds. Linear mixture analysis of planetary surface trates upon the VNIR, where there is more available composition requires access to reflectance spectra of signal. Yet, a thin film (<~10 microns) in the labora- the candidate compounds. Nonlinear mixture analysis tory does not engender sufficient path length for the requires the real and imaginary indices of refraction weak VNIR absorptions to manifest. This is not a (optical constants), which may be estimated from re- problem for a planetary regolith several meters to flectance spectra, or derived from a combination of kilometers thick, but does present a challenge for labo- reflectance and transmittance measurements. -

Supplement of Atmospheric Chemistry, Sources and Sinks Of
Supplement of Atmos. Chem. Phys., 17, 8789–8804, 2017 https://doi.org/10.5194/acp-17-8789-2017-supplement © Author(s) 2017. This work is distributed under the Creative Commons Attribution 3.0 License. Supplement of Atmospheric chemistry, sources and sinks of carbon suboxide, C3O2 Stephan Keßel et al. Correspondence to: Jonathan Williams ([email protected]) The copyright of individual parts of the supplement might differ from the CC BY 3.0 License. 1. Theoretical studies of additional reactions in the O3-initiated oxidation of C3O2 As discussed in the main text, the primary ozonide (POZ) formed after O3 addition on carbon suboxide was found to decompose either to an OOCCO Criegee intermediate + CO2, or to a cyclic CO2 dimer INT1 + CO. The transition state for this later channel is intriguing, as its geometry resembles that expected for the formation of the OOCO Criegee intermediate (INT2, Figure S1) with coproduct moiety O=C=C=O readily falling apart to 2 CO. Minimum energy pathway (IRC) calculations, however, indicate that during the dissociation process, only one CO is formed, while the remaining OC(OO)CO moiety rearranges to INT1. Figure S2 shows an animation of this process. It is currently not clear if this reaction path is affected by the methodology used; conceivably, the post-TS pathway bifurcates towards the formation of the OOCO (INT2) carbonyl oxide. We have additionally characterized a Criegee intermediate INT3, where the cyclization process forms a ring structure with the C-O moiety rather than the C-O-O moiety of the original POZ cyclic trioxilane decomposition. -

COVALENT COMPOUNDS Created By: Julio Cesar Torres Orozco
NOMENCLATURE IONIC-COVALENT COMPOUNDS Created by: Julio Cesar Torres Orozco Background: Before there was an established naming (nomenclature) system for chemical compounds, chemists assigned compounds a specific name for reference. Table salt (NaCl), water (H2O) and ammonia (NH3) are some of the most common examples of this. There are two types of chemical compounds that are important in general chemistry: Ionic and Covalent compounds. This handout will aim to explain the nomenclature system there exists for these types of compounds. Ionic compounds: METAL + NON-METAL RULES: ! Name of ionic compounds is composed of the name of the positive ion (from the metal) and the name of the negative ion. Examples: NaBr Sodium bromide MgCl2 Magnesium chloride (NH4)2SO4 Ammonium sulfate ! It is important that we learn how to name monoatomic positive ions. These are some examples: Na+ sodium Zn2+ zinc Ca2+ calcium H+ hydrogen K+ potassium Sr2+ strontium ! When there are positive ions that have more than one oxidation state (number), as in the case of transition metals, we would have to indicate the charge of the ion in Roman numeral in parentheses (I,II,III,IV,V,VI,VI) after the name of the specific element. Examples: Fe2+ iron(II) Fe3+ iron (III) Sn2+ tin(II) Sn4+ tin(IV) Cu+ copper(I) Cu2+ copper(II) NOMENCLATURE IONIC-COVALENT COMPOUNDS Created by: Julio Cesar Torres Orozco ! Positive polyatomic ions have common names ending in suffix -onium Examples: + H3O Hydronium + NH4 Ammonium Now that we have covered positive monoatomic and polyatomic ions, let us look at the naming of negative ions. -

Electron Irradiation of Carbon Dioxide-Carbon Disulphide Ice Analog and Its Implication on the Identification of Carbon Disulphide on Moon
J. Chem. Sci. Vol. 128, No. 1, January 2016, pp. 159–164. c Indian Academy of Sciences. DOI 10.1007/s12039-015-0996-6 Electron irradiation of carbon dioxide-carbon disulphide ice analog and its implication on the identification of carbon disulphide on Moon B SIVARAMAN∗ Atomic, Molecular and Optical Physics Division, Physical Research Laboratory, Ahmedabad 380 009, India e-mail: [email protected] MS received 25 September 2014; revised 19 August 2015; accepted 7 September 2015 Abstract. Carbon dioxide (CO2) and carbon disulphide (CS2) molecular ice mixture was prepared under low temperature (85 K) astrochemical conditions. The icy mixture irradiated with keV electrons simulates the irradiation environment experienced by icy satellites and Interstellar Icy Mantles (IIM). Upon electron irradi- ation the chemical composition was found to have altered and the new products from irradiation were found to be carbonyl sulphide (OCS), sulphur dioxide (SO2), ozone (O3), carbon trioxide (CO3), sulphur trioxide (SO3), carbon subsulphide (C3S2) and carbon monoxide (CO). Results obtained confirm the presence of CS2 molecules in lunar south-pole probed by the Moon Impact Probe (MIP). Keywords. Interstellar medium; interstellar icy mantles; icy satellites; radiation processing; carbon dioxide; carbon disulphide. 1. Introduction implantation on CO2 ices. However, proton implanta- tiononSO2 could not synthesize molecules containing 6 Icy satellites and Interstellar Icy Mantles (IIM) are H-S bonds. Electron irradiation of CS2 and O2 ices car- known to synthesize and harbour variety of molecules ried out at 12 K7 had shown several products bearing ranging from diatomic to complex molecules that take C-O and S-O bonds.