Functional Characterization of a Synthetic Hydrophilic Antifungal Peptide Derived from the Marine Snail Cenchritis Muricatus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Systematics and Ecology of the Mangrove-Dwelling Littoraria Species (Gastropoda: Littorinidae) in the Indo-Pacific
ResearchOnline@JCU This file is part of the following reference: Reid, David Gordon (1984) The systematics and ecology of the mangrove-dwelling Littoraria species (Gastropoda: Littorinidae) in the Indo-Pacific. PhD thesis, James Cook University. Access to this file is available from: http://eprints.jcu.edu.au/24120/ The author has certified to JCU that they have made a reasonable effort to gain permission and acknowledge the owner of any third party copyright material included in this document. If you believe that this is not the case, please contact [email protected] and quote http://eprints.jcu.edu.au/24120/ THE SYSTEMATICS AND ECOLOGY OF THE MANGROVE-DWELLING LITTORARIA SPECIES (GASTROPODA: LITTORINIDAE) IN THE INDO-PACIFIC VOLUME I Thesis submitted by David Gordon REID MA (Cantab.) in May 1984 . for the Degree of Doctor of Philosophy in the Department of Zoology at James Cook University of North Queensland STATEMENT ON ACCESS I, the undersigned, the author of this thesis, understand that the following restriction placed by me on access to this thesis will not extend beyond three years from the date on which the thesis is submitted to the University. I wish to place restriction on access to this thesis as follows: Access not to be permitted for a period of 3 years. After this period has elapsed I understand that James Cook. University of North Queensland will make it available for use within the University Library and, by microfilm or other photographic means, allow access to users in other approved libraries. All uses consulting this thesis will have to sign the following statement: 'In consulting this thesis I agree not to copy or closely paraphrase it in whole or in part without the written consent of the author; and to make proper written acknowledgement for any assistance which I have obtained from it.' David G. -

Malacofauna Marina Del Parque Nacional “Los Caimanes”, Villa Clara, Cuba
Tesis de Diploma Malacofauna Marina del Parque Nacional “Los Caimanes”, Villa Clara, Cuba. Autora: Liliana Olga Quesada Pérez Junio, 2011 Universidad Central “Marta Abreu” de Las Villas Facultad Ciencias Agropecuaria TESIS DE DIPLOMA Malacofauna marina del Parque Nacional “Los Caimanes”, Villa Clara, Cuba. Autora: Liliana Olga Quesada Pérez Tutor: M. C. Ángel Quirós Espinosa Investigador Auxiliar y Profesor Auxiliar [email protected] Centro de Estudios y Servicios Ambientales, CITMA-Villa Clara Carretera Central 716, Santa Clara Consultante: Dr.C. José Espinosa Sáez Investigador Titular Instituto de Oceanología Junio, 2011 Pensamiento “La diferencia entre una mala observación y una buena, es que la primera es errónea y la segunda es incompleta.” Van Hise Dedicatoria Dedicatoria: A mis padres, a Yandy y a mi familia: por las innumerables razones que me dan para vivir, y por ser fuente de inspiración para mis metas. Agradecimientos Agradecimientos: Muchos son los que de alguna forma contribuyeron a la realización de este trabajo, todos saben cuánto les agradezco: Primero quiero agradecer a mis padres, que aunque no estén presentes sé que de una forma u otra siempre estuvieron allí para darme todo su amor y apoyo. A mi familia en general: a mi abuela, hermano, a mis tíos por toda su ayuda y comprensión. A Yandy y a su familia que han estado allí frente a mis dificultades. Agradecer a mi tutor el M.Sc. Ángel Quirós, a mi consultante el Dr.C. José Espinosa y a la Dra.C. María Elena, por su dedicación para el logro de esta tesis. A mis compañeros de grupo por estos cinco años que hemos compartidos juntos, que para mí fueron inolvidables. -

Resistant Pseudosuccinea Columella Snails to Fasciola Hepatica (Trematoda) Infection in Cuba : Ecological, Molecular and Phenotypical Aspects Annia Alba Menendez
Comparative biology of susceptible and naturally- resistant Pseudosuccinea columella snails to Fasciola hepatica (Trematoda) infection in Cuba : ecological, molecular and phenotypical aspects Annia Alba Menendez To cite this version: Annia Alba Menendez. Comparative biology of susceptible and naturally- resistant Pseudosuccinea columella snails to Fasciola hepatica (Trematoda) infection in Cuba : ecological, molecular and phe- notypical aspects. Parasitology. Université de Perpignan; Instituto Pedro Kouri (La Havane, Cuba), 2018. English. NNT : 2018PERP0055. tel-02133876 HAL Id: tel-02133876 https://tel.archives-ouvertes.fr/tel-02133876 Submitted on 20 May 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Délivré par UNIVERSITE DE PERPIGNAN VIA DOMITIA En co-tutelle avec Instituto “Pedro Kourí” de Medicina Tropical Préparée au sein de l’ED305 Energie Environnement Et des unités de recherche : IHPE UMR 5244 / Laboratorio de Malacología Spécialité : Biologie Présentée par Annia ALBA MENENDEZ Comparative biology of susceptible and naturally- resistant Pseudosuccinea columella snails to Fasciola hepatica (Trematoda) infection in Cuba: ecological, molecular and phenotypical aspects Soutenue le 12 décembre 2018 devant le jury composé de Mme. Christine COUSTAU, Rapporteur Directeur de Recherche CNRS, INRA Sophia Antipolis M. Philippe JARNE, Rapporteur Directeur de recherche CNRS, CEFE, Montpellier Mme. -

UC Davis UC Davis Previously Published Works
UC Davis UC Davis Previously Published Works Title Molluscan marginalia: Serration at the lip edge in gastropods Permalink https://escholarship.org/uc/item/2mx5c6w9 Journal Journal of Molluscan Studies, 80(3) ISSN 0260-1230 Author Vermeij, GJ Publication Date 2014 DOI 10.1093/mollus/eyu020 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Journal of The Malacological Society of London Molluscan Studies Journal of Molluscan Studies (2014) 80: 326–336. doi:10.1093/mollus/eyu020 Advance Access publication date: 16 April 2014 Molluscan marginalia: serration at the lip edge in gastropods Geerat J. Vermeij Geology Department, University of California, One Shields Avenue, Davis, CA 95616, USA Correspondence: G.J. Vermeij; e-mail: [email protected] Downloaded from (Received 5 September 2013; accepted 10 February 2014) ABSTRACT The shells of many marine gastropods have ventrally directed serrations (serial projections) at the edge http://mollus.oxfordjournals.org/ of the adult outer lip. These poorly studied projections arise as extensions either of external spiral cords or of interspaces between cords. This paper describes taxonomic, phylogenetic, architectural and func- tional aspects of serrations. Cord-associated serrations occur in cerithiids, strombids, the personid Distorsio anus, ocenebrine muricids and some cancellariids. Interspace-associated serrations are phylo- genetically much more widespread, and occur in at least 16 family-level groups. The nature of serration may be taxonomically informative in some fissurellids, littorinids, strombids and costellariids, among other groups. Serrated outer lips occur only in gastropods in which the apex points more backward than upward, but the presence of serrations is not a necessary byproduct of the formation of spiral sculp- tural elements. -

228100 N, Tomando El Camino De Los Conucos Hasta El Entronque
228100 N, tomando el camino de los Conucos hasta el entronque de Zapato en 101700 E, 231400 N, siguiendo por el camino de Zapato hasta el final en 100700 E, 231600 N, girando al N hasta el estero de Santa Catalina por el cual sale al mar en 100300 E, 234950 N, se continúa por la línea de costa hasta el nacimiento del estero Los Morros en 096800 E, 234900 N, el cual se dirige hasta el punto N de Barra Sorda en 094300 E, 235400 N, por la orilla NO de esta barra en su límite con los manglares hasta cruzar el camino de Faro Roncali al estero Palmarito en 091900 E, 231900 N, en dirección S a 150 m del terraplén del Sur hasta el Pantano de Caleta Larga en 092800 E, 229950 N, donde bordea el Pantano hasta 092600 E, 229850 N, siguiendo la línea recta virtual hasta 090800 E, 228750 N, punto inicial de este derrotero. • A los efectos de controlar adecuadamente las acciones que puedan repercutir negativamente sobre esta área protegida se establece una Zona de Amortiguamiento que comprende los 500 metros a partir del límite externo del área y que se indica en el Anexo Cartográfico. Anexo 3 Listado de especies de la flora Flora terrestre Flora marina División RHODOPHYTA Orden CORALLINALES Familia CORALLINACEA 1. Amphiroa beauvoisii Lamouroux 2. Amphiroa fragilissima (Linnaeus) Lamouroux 3. Amphiroa rigida Lamouroux 4. Haliptilon cubense (Montagne ex Kützing) Garbary et Johansen 5. Haliptilon subulatum (Ellis et Solander) Johansen 6. Hydrolithon farinosum (Lamouroux) Penrose et Chamberlain 7. Jania adhaerens Lamouroux 8. -
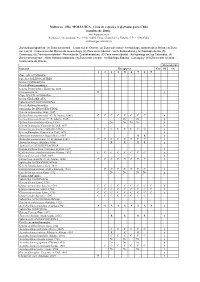
Moluscos - Filo MOLLUSCA
Moluscos - Filo MOLLUSCA. Lista de especies registradas para Cuba (octubre de 2006). José Espinosa Sáez Instituto de Oceanología, Ave 1ª No. 18406, Playa, Ciudad de La Habana, C.P. 11200, Cuba [email protected] Zonas biogeográficas: (1) Zona suroriental – Costa sur de Oriente, (2) Zona surcentral - Archipiélago Jardines de la Reina, (3) Zona sur central - Costa al sur del Macizo de Guamuhaya, (4) Zona suroccidental - Golfo de Batabanó y Archipiélago de los, (5) Canarreos, (6) Zona suroccidental - Península de Guanahacabibes, (7) Zona noroccidental - Archipiélago de Los Colorados, (8) Zona noroccidental - Norte Habana-Matanzas, (9) Zona norte-central - Archipiélago Sabana - Camagüey, (10) Zona norte-oriental - Costa norte de Oriente Abreviaturas Especies Bioegiones Cu Pl Oc 1 2 3 4 5 6 7 8 9 Clase APLACOPHORA Subclase SOLENOGASTRES Orden CAVIBELONIA Familia Proneomeniidae Género Proneomenia Hubrecht, 1880 Proneomenia sp . R x Clase POLYPLACOPHORA Orden NEOLORICATA Suborden ISCHNOCHITONINA Familia Ischnochitonidae Subfamilia ISCHNOCHITONINAE Género Ischnochiton Gray, 1847 Ischnochiton erythronotus (C. B. Adams, 1845) C C C C C C C C x Ischnochiton papillosus (C. B. Adams, 1845) Nc Nc x Ischnochiton striolatus (Gray, 1828) Nc Nc Nc Nc x Género Ischnoplax Carpenter in Dall, 1879 x Ischnoplax pectinatus (Sowerby, 1832) C C C C C C C C x Género Stenoplax Carpenter in Dall, 1879 x Stenoplax bahamensis Kaas y Belle, 1987 R R x Stenoplax purpurascens (C. B. Adams, 1845) C C C C C C C C x Stenoplax boogii (Haddon, 1886) R R R R x Subfamilia CALLISTOPLACINAE Género Callistochiton Carpenter in Dall, 1879 x Callistochiton shuttleworthianus Pilsbry, 1893 C C C C C C C C x Género Ceratozona Dall, 1882 x Ceratozona squalida (C. -

Temperature and Relative Humidity Effects on Water Loss and Hemolymph Osmolality of Littoraria Angulifera (Lamarck, 1822)
University of Texas Rio Grande Valley ScholarWorks @ UTRGV UTB/UTPA Electronic Theses and Dissertations Legacy Institution Collections 4-2014 Temperature and relative humidity effects on water loss and hemolymph osmolality of Littoraria angulifera (Lamarck, 1822) Phillip J. Rose The University of Texas Rio Grande Valley Follow this and additional works at: https://scholarworks.utrgv.edu/leg_etd Part of the Animal Sciences Commons, Environmental Sciences Commons, and the Oceanography and Atmospheric Sciences and Meteorology Commons Recommended Citation Rose, Phillip J., "Temperature and relative humidity effects on water loss and hemolymph osmolality of Littoraria angulifera (Lamarck, 1822)" (2014). UTB/UTPA Electronic Theses and Dissertations. 39. https://scholarworks.utrgv.edu/leg_etd/39 This Thesis is brought to you for free and open access by the Legacy Institution Collections at ScholarWorks @ UTRGV. It has been accepted for inclusion in UTB/UTPA Electronic Theses and Dissertations by an authorized administrator of ScholarWorks @ UTRGV. For more information, please contact [email protected], [email protected]. Temperature and Relative Humidity Effects on Water Loss and Hemolymph Osmolality of Littoraria angulifera (Lamarck, 1822) A Thesis Presented to the Faculty of the College of Science, Mathematics and Technology University of Texas at Brownsville In Partial Fulfillment of the Requirements for the Degree Master of Science In the field of Biology by Phillip J. Rose April 2014 Copyright By Phillip J. Rose April 2014 Acknowledgements I would like to acknowledge and especially thank the many people who assisted and/or contributed to this project in some way, shape, or form…………..and there were many! First, I would like to say a big thank you to the thesis committee comprised of Dr. -

Gastropod Molluscs of the Southern Area Of
PAIDEIA XXI Vol. 10, Nº 2, Lima, julio-diciembre 2020, pp. 289-310 ISSN Versión Impresa: 2221-7770; ISSN Versión Electrónica: 2519-5700 http://revistas.urp.edu.pe/index.php/Paideia ORIGINAL ARTICLE / ARTÍCULO ORIGINAL GASTROPOD MOLLUSCS OF THE SOUTHERN AREA OF CIENFUEGOS, FROM THE BEACH RANCHO LUNA TO THE MOUTH OF THE ARIMAO RIVER, CUBA MOLUSCOS GASTRÓPODOS DE LA ZONA SUR DE CIENFUEGOS, DESDE PLAYA RANCHO LUNA HASTA LA DESEMBOCADURA DEL RÍO ARIMAO, CUBA Oneida Calzadilla-Milian1*; Rafael Armiñana-García2,*; José Alexis Sarría- Martínez1; Rigoberto Fimia-Duarte3; Jose Iannacone4,5; Raiden Grandía- Guzmán6 & Yolepsy Castillo-Fleites2 1* Universidad de Cienfuegos «Carlos Rafael Rodríguez», Cienfuegos, Cuba. E-mail: ocalzadilla@ ucf.edu.cu; [email protected] 2 Universidad Central «Marta Abreu» de Las Villas, Villa Clara, Cuba. E-mail: rarminana@uclv. cu / ycfl [email protected] 3 Facultad de Tecnología de la Salud y Enfermería (FTSE). Universidad de Ciencias Médicas de Villa Clara (UCM-VC), Cuba. E-mail: rigoberto.fi [email protected] 4 Laboratorio de Ecología y Biodiversidad Animal (LEBA). Facultad de Ciencias Naturales y Matemáticas (FCNNM). Universidad Nacional Federico Villarreal (UNFV). Lima, Perú. 5 Facultad de Ciencias Biológicas. Universidad Ricardo Palma (URP). Lima, Perú. E-mail: [email protected] 6 Centro Nacional para la Producción de Animales de Laboratorio (CENPALAB), La Habana, Cuba. E-mail: [email protected] * Author for correspondence: [email protected] ABSTRACT The research presented shows a malacological survey of Cienfuegos' southern area, from “Rancho Luna” beach to the mouth of the “Arimao River”. The malacological studies ranged from January 2018 to December of the same year. -

44-Sep-2016.Pdf
Page 2 Vol. 44, No. 3 In 1972, a group of shell collectors saw the need for a national organization devoted to the interests of shell collec- tors; to the beauty of shells, to their scientific aspects, and to the collecting and preservation of mollusks. This was the start of COA. Our member- AMERICAN CONCHOLOGIST, the official publication of the Conchol- ship includes novices, advanced collectors, scientists, and shell dealers ogists of America, Inc., and issued as part of membership dues, is published from around the world. In 1995, COA adopted a conservation resolution: quarterly in March, June, September, and December, printed by JOHNSON Whereas there are an estimated 100,000 species of living mollusks, many PRESS OF AMERICA, INC. (JPA), 800 N. Court St., P.O. Box 592, Pontiac, IL 61764. All correspondence should go to the Editor. ISSN 1072-2440. of great economic, ecological, and cultural importance to humans and Articles in AMERICAN CONCHOLOGIST may be reproduced with whereas habitat destruction and commercial fisheries have had serious ef- proper credit. We solicit comments, letters, and articles of interest to shell fects on mollusk populations worldwide, and whereas modern conchology collectors, subject to editing. Opinions expressed in “signed” articles are continues the tradition of amateur naturalists exploring and documenting those of the authors, and are not necessarily the opinions of Conchologists the natural world, be it resolved that the Conchologists of America endors- of America. All correspondence pertaining to articles published herein es responsible scientific collecting as a means of monitoring the status of or generated by reproduction of said articles should be directed to the Edi- mollusk species and populations and promoting informed decision making tor. -

Mitochondrial DNA Hyperdiversity and Population Genetics in the Periwinkle Melarhaphe Neritoides (Mollusca: Gastropoda)
Mitochondrial DNA hyperdiversity and population genetics in the periwinkle Melarhaphe neritoides (Mollusca: Gastropoda) Séverine Fourdrilis Université Libre de Bruxelles | Faculty of Sciences Royal Belgian Institute of Natural Sciences | Directorate Taxonomy & Phylogeny Thesis submitted in fulfilment of the requirements for the degree of Doctor (PhD) in Sciences, Biology Date of the public viva: 28 June 2017 © 2017 Fourdrilis S. ISBN: The research presented in this thesis was conducted at the Directorate Taxonomy and Phylogeny of the Royal Belgian Institute of Natural Sciences (RBINS), and in the Evolutionary Ecology Group of the Free University of Brussels (ULB), Brussels, Belgium. This research was funded by the Belgian federal Science Policy Office (BELSPO Action 1 MO/36/027). It was conducted in the context of the Research Foundation – Flanders (FWO) research community ‘‘Belgian Network for DNA barcoding’’ (W0.009.11N) and the Joint Experimental Molecular Unit at the RBINS. Please refer to this work as: Fourdrilis S (2017) Mitochondrial DNA hyperdiversity and population genetics in the periwinkle Melarhaphe neritoides (Linnaeus, 1758) (Mollusca: Gastropoda). PhD thesis, Free University of Brussels. ii PROMOTERS Prof. Dr. Thierry Backeljau (90 %, RBINS and University of Antwerp) Prof. Dr. Patrick Mardulyn (10 %, Free University of Brussels) EXAMINATION COMMITTEE Prof. Dr. Thierry Backeljau (RBINS and University of Antwerp) Prof. Dr. Sofie Derycke (RBINS and Ghent University) Prof. Dr. Jean-François Flot (Free University of Brussels) Prof. Dr. Marc Kochzius (Vrije Universiteit Brussel) Prof. Dr. Patrick Mardulyn (Free University of Brussels) Prof. Dr. Nausicaa Noret (Free University of Brussels) iii Acknowledgements Let’s be sincere. PhD is like heaven! You savour each morning this taste of paradise, going at work to work on your passion, science. -

<I>Cenchritis Muricatus</I>
BULLETIN OF MARINE SCIENCE, 84(3): 307–313, 2009 E FFECTS of DISTURBANCE on CENCHRITIS MURICATUS (BEADED PERIWINKLE) pOPULATIONS ON small islanDS in THE BAHAMAS Jonah Piovia-Scott ABSTRACT D isturbance can have multiple impacts on shoreline gastropods. This study compares populations of a common supralittoral snail on islands in exposed and protected areas; the former are subject to much more disturbance from wave ac- tion and storm surges. Cenchritis muricatus (Linnaeus, 1758) density was six times higher on protected islands than on exposed islands, representing 2.5 times more biomass. Contrary to expectation, individuals were larger on exposed islands than on protected islands (mean lengths were 26.1 mm and 20.0 mm, respectively); this difference was primarily explained by a significant negative relationship between body size and density coupled with the fact that exposed islands had lower densities. I suggest that periodic large disturbances and oceanographic processes associated with dispersal limit the abundance of C. muricatus on exposed islands, and that larger sizes on exposed islands were probably due to enhanced growth caused by reduced intraspecific competition. D isturbance can have profound impacts on ecological communities (Sousa, 1984; Pickett and White, 1985). These impacts have been particularly well-studied in shoreline ecosystems where disturbance, usually in the form of wave action, affects a wide variety of taxa (Dayton, 1971; Paine and Levin, 1981; Underwood, 1999; Walker et al., 2008). Gastropods are a common component of most shoreline ecosystems, and disturbance can affect gastropods both directly, by inhibiting settlement (Crisp, 1955; Sousa, 1979; Bushek, 1988; Pawlik and Butman, 1993) or dislodging individuals from the substrate (Boulding and van Alstyne, 1993; Trussell, 1997), and indirectly, by removing predators (Menge and Sutherland, 1976; Menge, 1978), competitors (Steffani and Branch, 2003a,b), or facilitators (Underwood, 1999). -

Aqueous Extracts of Marine Invertebrates from Cuba Coastline
Indian Journal of Natural Products and Resources Vol. 8(2), June 2017, pp. 107-119 Aqueous extracts of marine invertebrates from Cuba coastline display neutral aminopeptidase inhibitory activities and effects on cancer cells and Plasmodium falciparum parasites Isel Pascual Alonso1*, Lotfi Bounaadja2, Laura Sánchez1, Laura Rivera1, Céline Tarnus3, Marjorie Schmitt4, Gabriela Garcia1, Lisset Diaz1, Aida Hernandez-Zanuy5, Belinda Sánchez6 and Isabelle Florent2 1Center for Protein Studies, Faculty of Biology, University of Havana, Cuba 2Molécules de Communication et Adaptation des Microorganismes (MCAM, UMR 7245), Sorbonne, Universités, Muséum national d’Histoire naturelle, CNRS, CP52, 57 rue Cuvier 75005 Paris, France 3Laboratoire de Chimie Organique et Bioorganique – COB, 4Laboratoire de Chimie Moléculaire - CNRS UMR7509, Institut de Recherche Jean-Baptiste Donnet, 3 bis rue Alfred Werner - 68093 Mulhouse Cedex, France 5Institute of Oceanology, AMA, CITMA, Cuba 6Center of Molecular Immunology, BioCubaFarma, Cuba Received 15 September 2016; Revised 01March 2017 Neutral aminopeptidases are enzymes distributed in all living organisms. By hydrolyzing biologically active peptides in tissues and biological fluids, they are involved in the control of many physiological processes. They became established targets for new therapeutic agents in cancer, but also in parasitic diseases like malaria. Marine organisms are promising sources for biomolecules but few examples of neutral aminopeptidase inhibitors are described. The goal of this work was to search in Cuban marine invertebrates, for inhibitory activities of neutral aminopeptidases of biomedical relevance, belonging to the M1 and M17 metallopeptidase families. The screening of inhibitory activities was performed using aqueous crude extracts and their 2.5 % TCA treatments. The treatments with 2.5 % TCA increased the recovery of inhibitory activities versus all enzymes tested and from all of marine species.