Result of National Maths Olympiad 2018-19
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Secretariat Phones
H.P.SECRETARIAT TELEPHONE NUMBERS th AS ON 09 August, 2021 DESIGNATION NAME ROOM PHONE PBX PHONE NO OFFICE RESIDENCE HP Sectt Control Room No. E304-A 2622204 459,502 HP Sectt Fax No. 2621154 HP Sectt EPABX No. 2621804 HP Sectt DID Code 2880 COUNCIL OF MINISTERS CHIEF MINISTER Jai Ram Thakur 1st Floor 2625400 600 Ellerslie 2625819 Building 2620979 2627803 2627808 2628381 2627809 FAX 2625011 Oak-Over 858 2621384 2627529 FAX 2625255 While in Delhi Residence Office New Delhi Tel fax 23329678 Himachal Sadan 24105073 24101994 Himachal Bhawan 23321375 Advisor to CM + Dr.R.N.Batta 1 st Floor 2625400 646 2627219 Pr.PS.to CM Ellersile 2625819 94180-83222 PS to Pr.PS to CM Raman Kumar Sharma 2625400 646 2835180 94180-23124 OSD to Chief Minister Mahender Kumar Dharmani E121 2621007 610,643 2628319 94180-28319 PS to OSD Rajinder Verma E120 2621007 710 94593-93774 OSD to Chief Minister Shishu Dharma E16G 2621907 657 2628100 94184-01500 PS to OSD Balak Ram E15G 2621907 757 2830786 78319-80020 Pr. PS to Diwan Negi E102 2627803 799 2812250 Chief Minister 94180-20964 Sr.PS to Chief Minister Subhash Chauhan E101 2625819 785 2835863 98160-35863 Sr.PS to Chief Minister Satinder Kumar E101 2625819 743 2670136 94180-80136 PS to Chief Minister Kaur Singh Thakur E102 2627803 700 2628562 94182-32562 PS to Chief Minister Tulsi Ram Sharma E101 2625400 746,869 2624050 94184-60050 Press Secretary Dr.Rajesh Sharma E104 2620018 699 94180-09893 to C M Addl.SP. Brijesh Sood E105 2627811 859 94180-39449 (CM Security) CEO Rajeev Sharma 23 G & 24 G 538,597 94184-50005 MyGov. -

Lesson. 8 Devotional Paths to the Divine
Grade VII Lesson. 8 Devotional paths to the Divine History I Multiple choice questions 1. Religious biographies are called: a. Autobiography b. Photography c. Hierography d. Hagiography 2. Sufis were __________ mystics: a. Hindu b. Muslim c. Buddha d. None of these 3. Mirabai became the disciple of: a. Tulsidas b. Ravidas c. Narsi Mehta d. Surdas 4. Surdas was an ardent devotee of: a. Vishnu b. Krishna c. Shiva d. Durga 5. Baba Guru Nanak born at: a. Varanasi b. Talwandi c. Ajmer d. Agra 6. Whose songs become popular in Rajasthan and Gujarat? a. Surdas b. Tulsidas c. Guru Nanak d. Mira Bai 7. Vitthala is a form of: a. Shiva b. Vishnu c. Krishna d. Ganesha 8. Script introduced by Guru Nanak: a. Gurudwara b. Langar c. Gurmukhi d. None of these 9. The Islam scholar developed a holy law called: a. Shariat b. Jannat c. Haj d. Qayamat 10. As per the Islamic tradition the day of judgement is known as: a. Haj b. Mecca c. Jannat d. Qayamat 11. House of rest for travellers kept by a religious order is: a. Fable b. Sama c. Hospice d. Raqas 12. Tulsidas’s composition Ramcharitmanas is written in: a. Hindi b. Awadhi c. Sanskrit d. None of these 1 Created by Pinkz 13. The disciples in Sufi system were called: a. Shishya b. Nayanars c. Alvars d. Murids 14. Who rewrote the Gita in Marathi? a. Saint Janeshwara b. Chaitanya c. Virashaiva d. Basavanna 1. (d) 2. (b) 3. (b) 4. (b) 5. (b) 6. (d) 7. -

Supplementary Materials For
Supplementary Materials for Genomic characterization and Epidemiology of an emerging SARS-CoV-2 variant in Delhi, India Mahesh S Dhar#, Robin Marwal#, Radhakrishnan VS#, Kalaiarasan Ponnusamy#, Bani Jolly#, Rahul C. Bhoyar#, Viren Sardana, Salwa Naushin, Mercy Rophina, Thomas A Mellan, Swapnil Mishra, Charlie Whittaker, Saman Fatihi, Meena Datta, Priyanka Singh, Uma Sharma, Rajat Ujjainiya, Nitin Bhateja, Mohit Kumar Divakar, Manoj K Singh, Mohamed Imran, Vigneshwar Senthivel, Ranjeet Maurya, Neha Jha, Priyanka Mehta, Vivekanand A, Pooja Sharma, Arvinden VR, Urmila Chaudhary, Lipi Thukral, Seth Flaxman, Samir Bhatt, Rajesh Pandey, Debasis Dash, Mohammed Faruq, Hemlata Lall, Hema Gogia, Preeti Madan, Sanket Kulkarni, Himanshu Chauhan, Shantanu Sengupta, Sandhya Kabra, The Indian SARS-CoV-2 Genomics Consortium (INSACOG), Ravindra K. Gupta, Sujeet K Singh, Anurag Agrawal*, Partha Rakshit* #Equal contribution *Correspondence to: [email protected] (Partha Rakshit), [email protected] (Anurag Agrawal) This PDF file includes: Materials and Methods Figs. S1 to S3 Tables S1 References 1 Materials and Methods Genome Sequencing and Analysis Nasopharyngeal and throat swab samples from COVID-19 confirmed cases with Ct value < 25 was collected and transported to Biotechnology Division, NCDC from the various testing sites across the States as per the sampling strategy of Central Surveillance Unit (CSU) of Integrated Disease Surveillance Programme (IDSP), NCDC. Twenty-seven post-vaccinated COVID-19 positive samples were also included in this study. All patient details were filled on the patient identification form and were accompanied with the samples. A total of 11,335 samples were received for genomic sequencing at NCDC from Nov 2020 till May 2021. The RNA was isolated using MagNA Pure RNA extraction system (Roche). -

List of Candidates (Provisionally Eligiblenot Eligible) for Research
Central University of Rajasthan Ph.D. Admissions 2021 List of Candidates (Provisionally Eligible/Not Eligible) for Research Aptitude Test, Presentation and Interview Ph.D. in Atmospheric Sciences Provisionally ID No. Student_Name Eligible Remarks (YES / NO) 116 SUBHASH YADAV YES 1227 ANIRUDH SHARMA YES 1345 ROHITASH YADAV YES 219 RITU BHARGAVA RAI YES Subject to submission of application through proper channel 363 SURINA ROUTRAY YES 396 SHANTANU SINHA YES 404 SHRAVANI BANERJEE YES 915 RAJNI CHOUDHARY YES 956 SONIYA YADAV YES 425 JYOTI PATHAK YES 85 SURUCHI YES 1404 BARKHA KUMAWAT NO No proof of qualifying any National Level Examination 988 ANJANA YADAV NO No proof of National Level Examination NOTE: Grievances, if any, w.r.t. above list may be submitted on [email protected] on or before 11th August 2021. The above list is only to appear for Research Aptitude Test, Presentation and Interview and it does not give any claim of final Admission to Ph.D. programme. Page No. 1 of 31 Central University of Rajasthan Ph.D. Admissions 2021 List of Candidates (Provisionally Eligible/Not Eligible) for Research Aptitude Test, Presentation and Interview Ph.D. in Biochemistry Provisionally ID No. Student_Name Eligible Remarks (YES / NO) 1000 SIMRAN BANSAL YES 1032 RAJNI YES 1047 AKASH CHOUDHARY YES 1048 AKASH CHOUDHARY YES 1050 ADITI SHARMA YES 1065 NAVEEN SONI YES 1168 NIDHI KUMARI YES 1185 ANISHA YES 1206 ABHISHEK RAO YES 1212 RASHMI YES 1228 NEELAM KUMARI YES 1241 ANUKRATI ARYA YES 1319 -

Sikhism Reinterpreted: the Creation of Sikh Identity
Lake Forest College Lake Forest College Publications Senior Theses Student Publications 4-16-2014 Sikhism Reinterpreted: The rC eation of Sikh Identity Brittany Fay Puller Lake Forest College, [email protected] Follow this and additional works at: http://publications.lakeforest.edu/seniortheses Part of the Asian History Commons, History of Religion Commons, and the Religion Commons Recommended Citation Puller, Brittany Fay, "Sikhism Reinterpreted: The rC eation of Sikh Identity" (2014). Senior Theses. This Thesis is brought to you for free and open access by the Student Publications at Lake Forest College Publications. It has been accepted for inclusion in Senior Theses by an authorized administrator of Lake Forest College Publications. For more information, please contact [email protected]. Sikhism Reinterpreted: The rC eation of Sikh Identity Abstract The iS kh identity has been misinterpreted and redefined amidst the contemporary political inclinations of elitist Sikh organizations and the British census, which caused the revival and alteration of Sikh history. This thesis serves as a historical timeline of Punjab’s religious transitions, first identifying Sikhism’s emergence and pluralism among Bhakti Hinduism and Chishti Sufism, then analyzing the effects of Sikhism’s conduct codes in favor of militancy following the human Guruship’s termination, and finally recognizing the identity-driven politics of colonialism that led to the partition of Punjabi land and identity in 1947. Contemporary practices of ritualism within Hinduism, Chishti Sufism, and Sikhism were also explored through research at the Golden Temple, Gurudwara Tapiana Sahib Bhagat Namdevji, and Haider Shaikh dargah, which were found to share identical features of Punjabi religious worship tradition that dated back to their origins. -

Substantial and Substantive Corporeality in the Body Discourses of Bhakti Poets
Perichoresis Volume 18.2 (2020): 73–94 DOI: 10.2478/perc-2020-0012 SUBSTANTIAL AND SUBSTANTIVE CORPOREALITY IN THE BODY DISCOURSES OF BHAKTI POETS YADAV SUMATI* PG Govt. College for Girls, Chandigarh, India ABSTRACT. This paper studies the representation of human corporeal reality in the discours- es of selected Bhakti poets of the late medieval period in India. Considering the historical background of the Bhakti movement and contemporary cultural milieu in which these mystic poets lived, their unique appropriation of the ancient concept of body is reviewed as revolu- tionary. The focus of the study is the Kabir Bijak, Surdas’s Vinay-Patrika, and Tulsidas’s Vinay- Patrika, wherein they look at and beyond the organic corporeality and encounter human body not as a socially, religiously, economically stamped noble body or lowly body; male body or female body, but a human body. This paper explores how, like existential phenomenologists, these poet/singers decode the material reality of human beings and link it to the highest goal of achieving Moksha (liberation from the cycle of birth-death) by making body a vulnerable but essential instrument towards spiritual awakening. The paper also reflects upon how these poets have suggested a middle path of absolute devotion to God while performing all earthly duties, seek spiritual enlightenment and avoid the extremities of asceticism and hedonism. KEYWORDS: corporeality, body, liberation, salvation, bhakti In this Kali Yug the body is full of woe, care, wickedness and diverse pains. Where there is steadfastness, peace and all purity, rise, Kabir, and meet it there. (Kabir) Those powerful rulers who had conquered the whole world, even made Yamraj (the God of death) their captive and tied him up—even they became the food of Kaal (Time), what do you count then? Contemplate and think about the whole matter seriously yourself—what is the truth, what is the reality. -

Prof. J. N. Gautam)
Dr. Jagdish Narain Gautam School of Studies in Library and Information Science, Jiwaji University, Gwalior – 474011. Mob. +919425364178 PHONE NO.0751-2343176 (R) 0751-2442627 E-mail: [email protected] FATHER'S NAME : Late Shri Asha Ram Gautam SPECIALIZATION : Knowledge Organization and Processing (Classification & Cataloguing), Library Information and Society, Library Management System and Library Automation and Networking. PRESENT POSITION : Professor, SOS, Library and Information Science. Rector, Jiwaji University, Gwalior. OTHER RESPONSIBILITIES : : Dean Faculty of Arts, Since 2006 : Chairman Board of Studies in LIS, Since 1996 : Member of Academic Council : Member of Planning and Evaluation Board : Chief Warden since 2005-2009 ACADEMIC EXPERIENCE : 22 Years QUALIFICATIONS : Degree Year Board/University Subject/Title Division Ph.D. 1991 Jiwaji University, The Information needs and Information Gwalior seeking of Physics & Chemistry Teachers in University and Collages of Gwalior and Chambal Division – A Scientific Study M.Lib.I.Sc 1986 Dr. H. S. Gour Library and Information Science II University, Sagar M.Sc. (Phy. 1984 Dr. H. S. Gour Anthropology I Anthro.) University, Sagar PROFESSIONAL EXPERIENCE: 22 Years (As per the details below) S. No. Post Held Department/Institution Duration 1. Librarian (Part -Time) Lok Kala Academy, Sagar. 01.09.1984 to 01.08.1985 2. Librarian Govt. P.G. College, Tikamgarh. 03.08.1985 to 04.09.1985 3. Librarian Govt. Degree College, Sitapur. 04.10.1986 to 31.07.1987 4. Assistant Librarian Central Library, Jiwaji University, 20.11.1987 to 08.05.1996 Gwalior. 5. In charge University Central Library, Jiwaji University, 09.05.1996 to 31.03.2010 Librarian Gwalior. TEACHING EXPERIENCE : 22 Years (As per the details below) B.Lib.I.Sc: Papers Taught - Knowledge organization and Processing (Classification & Cataloguing Theory) - Knowledge organization and Processing (Classification Practical) CC Rev. -
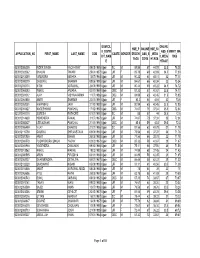
Application No First Name Last Name Dob Domicil
DOMICIL ONLINE HSE_P ONLINE HSE_50 E_DISTR _ASS_5 MERIT_MA APPLICATION_NO FIRST_NAME LAST_NAME DOB CASTE GENDER ERCEN _ASS_S _WEIG ICT_NAM 0_WEIG RKS TAGE CORE HTAGE E HTAGE 030812056209 INDER SINGH KACHHWAY 09/05/1986 Ujjain SC M 89.56 67 44.78 33.5 78.28 030812023062 SHALINI TIWARI 29/04/1987 Ujjain UR F 85.78 69 42.89 34.5 77.39 030812013389 VIRENDRA MISHRA 13/07/1984 Ujjain UR M 90.22 64 45.11 32 77.11 030812093079 DHEERAJ SHARMA 08/06/1990 Ujjain UR M 84.67 66 42.34 33 75.34 030812032174 NITIN AGRAWAL 26/09/1989 Ujjain UR M 80.44 69 40.22 34.5 74.72 030812068283 RAHUL ANJANA 02/11/1989 Ujjain OBC M 81.33 67 40.67 33.5 74.17 030812001012 AJAY VISHWAKARMA 11/07/1986 Ujjain OBC M 84.89 63 42.45 31.5 73.95 030812061800 AARTI SHARMA 22/01/1991 Ujjain UR F 83.2 64 41.6 32 73.6 030812053051 HIMANSHU JAIN 31/10/1988 Ujjain UR M 80.89 65 40.45 32.5 72.95 030312081427 NAGESHWAR PANCHAL 17/12/1989 Ujjain OBC M 74.67 71 37.34 35.5 72.84 030312041110 SURESH RATHORE 01/07/1991 Ujjain SC M 88 57 44 28.5 72.5 030812014630 HEMENDRA RAWAL 31/07/1987 Ujjain UR M 74.67 70 37.34 35 72.34 030812055427 LEELADHAR PANCHAL 21/11/1991 Ujjain OBC M 85.6 59 42.8 29.5 72.3 030812086499 VIJAY DHAKITE 21/01/1986 Ujjain SC M 81.56 62 40.78 31 71.78 030812115790 GAURAV SHRIVASTAVA 08/06/1986 Ujjain UR M 75.56 68 37.78 34 71.78 030812020950 ANKIT BIHANI 25/05/1988 Ujjain UR M 77.56 66 38.78 33 71.78 030812069570 PUSHPENDRA SINGH YADAV 02/03/1987 Ujjain OBC M 85.33 58 42.67 29 71.67 030812041960 YOGENDRA CHAUHAN 09/10/1986 Ujjain UR M 75.11 68 37.56 34 71.56 030812013862 -

Namdev Life and Philosophy Namdev Life and Philosophy
NAMDEV LIFE AND PHILOSOPHY NAMDEV LIFE AND PHILOSOPHY PRABHAKAR MACHWE PUBLICATION BUREAU PUNJABI UNIVERSITY, PATIALA © Punjabi University, Patiala 1990 Second Edition : 1100 Price : 45/- Published by sardar Tirath Singh, LL.M., Registrar Punjabi University, Patiala and printed at the Secular Printers, Namdar Khan Road, Patiala ACKNOWLEDGEMENTS I am grateful to the Punjabi University, Patiala which prompted me to summarize in tbis monograpb my readings of Namdev'\i works in original Marathi and books about him in Marathi. Hindi, Panjabi, Gujarati and English. I am also grateful to Sri Y. M. Muley, Director of the National Library, Calcutta who permitted me to use many rare books and editions of Namdev's works. I bave also used the unpubIi~bed thesis in Marathi on Namdev by Dr B. M. Mundi. I bave relied for my 0pIDlOns on the writings of great thinkers and historians of literature like tbe late Dr R. D. Ranade, Bhave, Ajgaonkar and the first biographer of Namdev, Muley. Books in Hindi by Rabul Sankritya)'an, Dr Barathwal, Dr Hazariprasad Dwivedi, Dr Rangeya Ragbav and Dr Rajnarain Maurya have been my guides in matters of Nath Panth and the language of the poets of this age. I have attempted literal translations of more than seventy padas of Namdev. A detailed bibliography is also given at the end. I am very much ol::lig(d to Sri l'and Kumar Shukla wbo typed tbe manuscript. Let me add at the end tbat my family-god is Vitthal of Pandbarpur, and wbat I learnt most about His worship was from my mother, who left me fifteen years ago. -
Bani of Bhagats-Part II.Pmd
BANI OF BHAGATS Complete Bani of Bhagats as enshrined in Shri Guru Granth Sahib Part II All Saints Except Swami Rama Nand And Saint Kabir Ji Dr. G.S. Chauhan Publisher : Dr. Inderjit Kaur President All India Pingalwara Charitable Society (Regd.) Amritsar-143001 Website:www.pingalwara.co; E-mail:[email protected] BANI OF BHAGATS PART : II Author : G.S. Chauhan B-202, Shri Ganesh Apptts., Plot No. 12-B, Sector : 7, Dwarka, New Delhi - 110075 First Edition : May 2014, 2000 Copies Publisher : Dr. Inderjit Kaur President All India Pingalwara Charitable Society (Regd.) Amritsar-143001 Ph : 0183-2584586, 2584713 Website:www.pingalwara.co E-mail:[email protected] (Link to download this book from internet is: pingalwara.co/awareness/publications-events/downloads/) (Free of Cost) Printer : Printwell 146, Industrial Focal Point, Amritsar Dedicated to the sacred memory of Sri Guru Arjan Dev Ji Who, while compiling bani of the Sikh Gurus, included bani of 15 saints also, belonging to different religions, castes, parts and regions of India. This has transformed Sri Guru Granth Sahib from being the holy scripture of the Sikhs only to A Unique Universal Teacher iii Contentsss • Ch. 1: Saint Ravidas Ji .......................................... 1 • Ch. 2: Sheikh Farid Ji .......................................... 63 • Ch. 3: Saint Namdev Ji ...................................... 113 • Ch. 4: Saint Jaidev Ji......................................... 208 • Ch. 5: Saint Trilochan Ji .................................... 215 • Ch. 6: Saint Sadhna Ji ....................................... 223 • Ch. 7: Saint Sain Ji ............................................ 227 • Ch. 8: Saint Peepa Ji.......................................... 230 • Ch. 9: Saint Dhanna Ji ...................................... 233 • Ch. 10: Saint Surdas Ji ...................................... 240 • Ch. 11: Saint Parmanand Ji .............................. 244 • Ch. 12: Saint Bheekhan Ji................................ -

Historical Perspective of Rajput Society
SOCIO - HISTORICAL PERSPECTIVE OF RAJPUT SOCIETY DR. YASHPALSINH V. RATHOD At. Dhamboliya Dist. Arvvali (GJ) INDIA Maharaja Shree Ranjorsinh has identified Kshatriya Pedigree, as per him 16 from Sun, 4 from Moon, 2 from Nagvanshi, 3 from Rushivanshi and 11 from Agnivanshi are also Kshatriya. There are some assumptions for 36 Descent. But in sun vansh Rathod, Katchvaha, Sisodiya, Badgujar,Kathariya, Sikarval, Nikumbh and Rekvar are considered. European historian Easterson, Indian veda and cultural tradion indicates Rajputs are ancient Aaryajati. These gens are from Rajput and they had ruled India since vaidik period. That cast has provided bravely protection to our country, religion and culture. INTRODUCTION This article shows origin of Rajput society and their social, cultural, geographical, Economical and historical aspects. People from same cast who are living in different areas of Gujarat should know their Ancestor, their detail information is necessary today. Today’s young generation does not have proper information regarding their Descent.they should know their gotra and sub cast properly. They don’t believe in relation of brotherhood. If we ask question to any rauput youngster regarding their Descent and sub branch, that answer is not satisfactory. That’s why origin and historical detail study of Rathod Descent is required. Here detail information written of 36 Descent and their sub branch indicated in this article. Rathod Descent community is in majority in some areas of Gujarat, also in some part of the state there are Solanki, Chauhan, Parmar, Zala,Waghela etc… community are living. Kulgury, Bhat, Charan, Vahivarcha, Rani, Maga, Rao, Dhol community has played an important role in Indian Kshatriya gaurav gatha. -

Saints, Hagiographers, and Religious Experience: the Case of Tukaram and Mahipati
religions Article Saints, Hagiographers, and Religious Experience: The Case of Tukaram and Mahipati J. E. Llewellyn Department of Religious Studies, Missouri State University, 901 South National Avenue, Springfield, MO 65897, USA; [email protected] Received: 27 December 2018; Accepted: 12 February 2019; Published: 15 February 2019 Abstract: One of the most important developments in Hinduism in the Common Era has been the rise of devotionalism or bhakti. Though theologians and others have contributed to this development, the primary motive force behind it has been poets, who have composed songs celebrating their love for God, and sometimes lamenting their distance from Her. From early in their history, bhakti traditions have praised not only the various gods, but also the devotional poets as well. And so hagiographies have been written about the lives of those exceptional devotees. It could be argued that we find the religious experience of these devotees in their own compositions and in these hagiographies. This article will raise questions about the reliability of our access to the poets’ religious experience through these sources, taking as a test case the seventeenth century devotional poet Tukaram and the hagiographer Mahipati. Tukaram is a particularly apt case for a study of devotional poetry and hagiography as the means to access the religious experience of a Hindu saint, since scholars have argued that his works are unusual in the degree to which he reflects on his own life. We will see why, for reasons of textual history, and for more theoretical reasons, the experience of saints such as Tukaram must remain elusive.