Supplementary Information for Changes in the Plasmodesma Structure and Permeability at the Bundle Sheath and Mesophyll Interface
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diversity of Plant Virus Movement Proteins: What Do They Have in Common?
processes Review Diversity of Plant Virus Movement Proteins: What Do They Have in Common? Yuri L. Dorokhov 1,2,* , Ekaterina V. Sheshukova 1, Tatiana E. Byalik 3 and Tatiana V. Komarova 1,2 1 Vavilov Institute of General Genetics Russian Academy of Sciences, 119991 Moscow, Russia; [email protected] (E.V.S.); [email protected] (T.V.K.) 2 Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia 3 Department of Oncology, I.M. Sechenov First Moscow State Medical University, 119991 Moscow, Russia; [email protected] * Correspondence: [email protected] Received: 11 November 2020; Accepted: 24 November 2020; Published: 26 November 2020 Abstract: The modern view of the mechanism of intercellular movement of viruses is based largely on data from the study of the tobacco mosaic virus (TMV) 30-kDa movement protein (MP). The discovered properties and abilities of TMV MP, namely, (a) in vitro binding of single-stranded RNA in a non-sequence-specific manner, (b) participation in the intracellular trafficking of genomic RNA to the plasmodesmata (Pd), and (c) localization in Pd and enhancement of Pd permeability, have been used as a reference in the search and analysis of candidate proteins from other plant viruses. Nevertheless, although almost four decades have passed since the introduction of the term “movement protein” into scientific circulation, the mechanism underlying its function remains unclear. It is unclear why, despite the absence of homology, different MPs are able to functionally replace each other in trans-complementation tests. Here, we consider the complexity and contradictions of the approaches for assessment of the ability of plant viral proteins to perform their movement function. -
Chapter 4 – Cell Structure
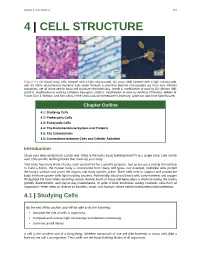
Chapter 4 | Cell Structure 107 4 | CELL STRUCTURE Figure 4.1 (a) Nasal sinus cells (viewed with a light microscope), (b) onion cells (viewed with a light microscope), and (c) Vibrio tasmaniensis bacterial cells (seen through a scanning electron microscope) are from very different organisms, yet all share certain basic cell structure characteristics. (credit a: modification of work by Ed Uthman, MD; credit b: modification of work by Umberto Salvagnin; credit c: modification of work by Anthony D'Onofrio, William H. Fowle, Eric J. Stewart, and Kim Lewis of the Lewis Lab at Northeastern University; scale-bar data from Matt Russell) Chapter Outline 4.1: Studying Cells 4.2: Prokaryotic Cells 4.3: Eukaryotic Cells 4.4: The Endomembrane System and Proteins 4.5: The Cytoskeleton 4.6: Connections between Cells and Cellular Activities Introduction Close your eyes and picture a brick wall. What is the wall's basic building block? It is a single brick. Like a brick wall, cells are the building blocks that make up your body. Your body has many kinds of cells, each specialized for a specific purpose. Just as we use a variety of materials to build a home, the human body is constructed from many cell types. For example, epithelial cells protect the body's surface and cover the organs and body cavities within. Bone cells help to support and protect the body. Immune system cells fight invading bacteria. Additionally, blood and blood cells carry nutrients and oxygen throughout the body while removing carbon dioxide. Each of these cell types plays a vital role during the body's growth, development, and day-to-day maintenance. -

Development and Function of Plasmodesmata in Zygotes of Fucus Distichus
Botanica Marina 2015; 58(3): 229–238 Chikako Nagasato*, Makoto Terauchi, Atsuko Tanaka and Taizo Motomura Development and function of plasmodesmata in zygotes of Fucus distichus Abstract: Brown algae have plasmodesmata, tiny tubu- Introduction lar cytoplasmic channels connecting adjacent cells. The lumen of plasmodesmata is 10–20 nm wide, and it takes a Multicellular organisms such as animals, fungi, land simple form, without a desmotubule (the inner membrane plants, and brown algae have specific cellular connections. structure consisting of endoplasmic reticulum in the plas- These structures connect the cytoplasm of adjacent cells modesmata of green plants). In this study, we analyzed and provide a route for cell-cell communication. Green the ultrastructure and distribution of plasmodesmata plants, including land plants and certain species of green during development of Fucus distichus zygotes. The first algae (reviewed in Robards and Lucas 1990, Raven 1997), cytokinesis of zygotes in brown algae is not accompanied and brown algae (Bisalputra 1966) possess plasmodes- by plasmodesmata formation. As the germlings develop, mata. These are cytoplasmic canals that pass through the plasmodesmata are found in all septal cell walls, includ- septal cell wall and connect adjacent cells. Plasmodes- ing the first cell division plane. Plasmodesmata are formed mata in green plants and brown algae share similar char- de novo on the existing cell wall. Pit fields, which are clus- acteristics; however, plasmodesmata in brown algae lack ters of plasmodesmata, were observed in germlings with a desmotubule, a tubular strand of connecting endoplas- differentiated cell layers. Apart from the normal plas- mic reticulum (ER) that penetrates the plasmodesmata modesmata, these pit fields had branched plasmodes- of land plants (Terauchi et al. -

Plasmodesmata and the Control of Symplastic Transport
Blackwell Science, LtdOxford, UKPCEPlant, Cell and Environment0016-8025Blackwell Publishing Ltd 2002 26 Original Article Plant, Cell and Environment (2003) 26, 103–124 Plasmodesmata and the control of symplastic transport A. G. ROBERTS & K. J. OPARKA Scottish Crop Research Institute, Invergowrie, Dundee, DD2 5DA, UK ‘There are holes in the sky other cells, some degree of intercellular connection is main- where the rain gets in, tained. This plasmodesmal continuum that potentially but they’re ever so small exists throughout the whole plant is termed the symplast that’s why rain’s so thin’ (Münch 1930). However, the symplast is not the open con- Spike Milligan (1968) tinuum that Münch originally hypothesized, but is divided into functional domains, each tightly regulated by different In 1879 Eduard Tangle discovered cytoplasmic connections forms of plasmodesmata (Erwee & Goodwin 1985; Ehlers between cells in the cotyledons of Strychnos nuxvomica, & Kollmann 2001). Plasmodesmata are now thought of as which he interpreted to be protoplasmic contacts. This led fluid, dynamic structures that can be modified both struc- him to hypothesize that ‘the protoplasmic bodies . are turally and functionally to cope with the requirements of united by thin strands passing through connecting ducts in specific cells and tissues. the walls, which put the cells into connection with each other and so unite them to an entity of higher order’ (Carr 1976). This challenged the then current view that cells func- THE STRUCTURE OF PLASMODESMATA tioned as autonomous units. It was after much research in Based on structure, two basic types of plasmodesmata have many other species and cell types that Strasburger, in 1901, been characterized; simple and branched. -

Tobacco Mosaic Virus Movement Protein Associates with the Cytoskeleton in Tobacco Cells
The Plant Cell, Vol. 7, 2101-21 14, December 1995 O 1995 American Society of Plant Physiologists Tobacco Mosaic Virus Movement Protein Associates with the Cytoskeleton in Tobacco Cells 8. Gail McLean, John Zupan, and Patricia C. Zambryskil Department of Plant Biology, University of California-Berkeley, Berkeley, California 94720-3102 Tobacco mosaic virus movement protein P30 complexes with genomic viral RNA for transport through plasmodesmata, the plant intercellular connections. Although most research with P30 focuses on its targeting to and gating of plasmodes- mata, the mechanisms of P30 intracellularmovement to plasmodesmata have not been defined. To examine P30 intracellular localization, we used tobacco protoplasts, which lack plasmodesmata, for transfection with plasmids carrying P30 cod- ing sequences under a constitutive promoter and for infection with tobacco mosaic virus particles. In both systems, P30 appears as filaments that colocalize primarily with microtubules. To a lesser extent, P30 filaments colocalize with actin filaments, and in vitro experiments suggested that P30 can bind directly to actin and tubulin. This association of P30 with cytoskeletal elements may play a critical role in intracellular transport of the P30-vira1 RNA complex through the cytoplasm to and possibly through plasmodesmata. INTRODUCTION To establish a systemic infection, plant viruses must move from that the cytoskeleton acts as a trafficking system for intracel- the infection site to the rest of the plant. For many plant viruses, lular transport, translocating vesicles, organelles, protein, and a virus-encoded product, the movement protein, actively poten- even mRNA to specific cellular locations (Williamson, 1986; tiates viral cell-to-cell spread through plasmodesmata, the Vale, 1987; Dingwall, 1992; Singer, 1992; Wilhelm and Vale, cytoplasmic bridges that function as intercellular connections 1993; Bassell et al., 1994; Hesketh, 1994). -

Organelle–Nucleus Cross-Talk Regulates Plant Intercellular
Organelle–nucleus cross-talk regulates plant PNAS PLUS intercellular communication via plasmodesmata Tessa M. Burch-Smitha, Jacob O. Brunkarda, Yoon Gi Choib, and Patricia C. Zambryskia,1 aDepartment of Plant and Microbial Biology and bFunctional Genomics Laboratory, University of California, Berkeley, CA 94720 Contributed by Patricia C. Zambryski, October 19, 2011 (sent for review June 20, 2011) We use Arabidopsis thaliana embryogenesis as a model system by either depositing or removing callose from the cell walls sur- for studying intercellular transport via plasmodesmata (PD). A rounding PD openings. PD also contain cytoskeleton-associated forward genetic screen for altered PD transport identified in- proteins, including actin (7) and myosin (8). Proteins with potential creased size exclusion limit (ise) 1 and ise2 mutants with increased roles in signal transduction are also PD constituents, such as a intercellular transport of fluorescent 10-kDa tracers. Both ise1 and calcium kinase (9), calreticulin (10), a kinase that phosphorylates ise2 exhibit increased formation of twinned and branched PD. viral movement proteins (11), and membrane receptor-like pro- ISE1 encodes a mitochondrial DEAD-box RNA helicase, whereas teins [PD-localized proteins (PDLPs)] (12). Remorin, a lipid-raft ISE2 encodes a DEVH-type RNA helicase. Here, we show that protein, was also recently found to localize to PD (13). Several ISE2 foci are localized to the chloroplast stroma. Surprisingly, additional potential membrane-bound PD-localized proteins have ise1 ise2 plastid development is defective in both and mutant recently been reported (14). embryos. In an effort to understand how RNA helicases that lo- We conducted a genetic screen of embryo-lethal mutants to calize to different organelles have similar impacts on plastid and identify genes critical to regulating PD function (15). -

Plasmodesmata in Higher Plants
2 PLASMODESMATA IN HIGHER PLANTS A.W. ROBARDS 1 Department of Developmental Biology, Research School of Biological Sciences, The Australian National University, Box 475, P.O., Canberra City, A.C.T. 2601, Australia 2.1. INTRODUCTION The first detailed description of "offene Communiaationen", or protoplasmic connecting threads is usually attributed to Tang1 (1879) although other workers subsequently contested his precedence. However, Tang1 was the first botanist to write at length on these structures and his article stimulated a spate of publications wbich described such connections between cells from all parts of the plant kingdom (for fuller details refer to Meeuse, 1941b; 1957, and Chapter 14). In 1901 Strasburger used the term "PZasmodesmen" to describe the protoplasmic connections and, despite numerous other suggestions (see Meeuse, 1957), the word has survived the test of time and is now almost universally accepted (English - plasmodesma - Gk. pZasma, form; desma, bond - P1u. plasmodesmata). Virtually all the early investigations involved treatment of cells to cause swelling of the wall so that plasmodesmata could be demonstrated by optical microscopy. This led to many critic isms and, as recently as 1964, Livingston reconsidered the "nature of pZasmodesmata in normaZ (Ziving) pZant tissue", so highlighting the problem that has been with us for over 80 years. The advent of elec tron microscopy has done much to expand knowledge about the structure and variability of plasmodesmata; it has been of rather less value in helping us to understand the true nature and physiological function of these connections. In this Chapter I shall collate some of the information relating to the distribution, structure and possible phys iological roles of plasmodesmata. -

Changes in the Plasmodesma Structure and Permeability at the Bundle Sheath and Mesophyll Interface During the Maize C4 Leaf Development
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.30.320283; this version posted October 1, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Title: Changes in the plasmodesma structure and permeability at the bundle sheath and mesophyll interface during the maize C4 leaf development. Peng Gao1, Baijuan Du2, Pinghua Li2, and Byung-Ho Kang1* 1State Key Laboratory of Agrobiotechnology, Center for Cell and Developmental Biology, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong, China 2State Key Laboratory of Crop Biology, College of Agronomic Sciences, Shandong Agricultural University, Tai’an 271018, China *Author for correspondence: Byung-Ho Kang ([email protected], 852-3943-6101) Key Words: C4 photosynthesis, plasmodesmata, maize, Kranz anatomy, dimorphic chloroplast, suberin Author contributions: GP and BHK: Planned and designed the research. GP: Performed microscopy imaging, immunolocalization, and qRT-PCR. BD and PL: Maintained maize mutant lines, carried out transcriptomic data analysis and gene expression correlation test. GP, BD, PL, and BHK: Interpreted the data. GP and BHK: Wrote the manuscript. bioRxiv preprint doi: https://doi.org/10.1101/2020.09.30.320283; this version posted October 1, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Plasmodesmata are intercellular channels that facilitate molecular diffusion betWeen neighboring plant cells. The development and functions of plasmodesmata are controlled by multiple intra- and intercellular signaling pathways. Plasmodesmata are critical for dual-cell C4 photosynthesis in maize because plasmodesmata at the mesophyll and bundle sheath interface mediate exchange of CO2-carrying organic acids. -

Tissue Preparation and Substructure of Plasmodesmata
CHAPTER 3 Tissue Preparation and Substructure of Plasmodesmata B. DING Department of Botany, Oklahoma State University, Stillwater, Oklahoma 74078, USA 1 Introduction 38 2...., Structure of the Plasmodesma 38 3 Methodologies of Tissue Preparation for Studying the Plasmodesmal Structure 39 3.1 Chemical Fixation 39 3.2 CryofIxation 42 3.2.1 Freeze Fracture 43 3.2.2 Freeze Substitution 44 4 Elucidating the Molecular Structure of the Plasmodesma by Integrative Approaches 45 5 Concluding Remarks 47 References 47 A. J. E. van Bel et al. (eds.), Plasmodesmata © Springer-Verlag Berlin Heidelberg 1999 38 CHAPTER 3 Tissue Preparation and Substructure of Plasmodesmata 1 Introduction Research on the plasmodesma seeks to address two basic issues: what kinds of mole cules are transported through this intercellular organelle, and how are these molecules transported? Elucidation of the transport mechanisms requires that the substructure of the plasmodesma be understood. This understanding may include at least two as pects: ultrastructural features and molecular compositions. Views of the plasmodesmal substructure have mainly been obtained through the use of transmission electron microscopy (TEM). This chapter attempts to discuss in general terms the prevailing methods used to prepare tissue samples for TEM studies of plasmodesmata, focusing on the limitations of these methods and on the interpre tation of data obtained with these methods. I will also discuss briefly the use of inte grated approaches to study the plasmodesmal structure at the molecular level. 2 Structure of the Plasmodesma The structure of the plasmodesma is reviewed extensively in Chapter 9 and only some general features are outlined here to facilitate subsequent technical discussions. -

The Structure of Plasmodesmata As Revealed by Plasmolysis, Detergent Extraction, and Protease Digestion
The Structure of Plasmodesmata as Revealed by Plasmolysis, Detergent Extraction, and Protease Digestion Lewis G. T'flney,* Todd J. Cooke,~ Patricia S. Connelly,* and Mary S. Tilney* * Department of Biology, University of Pennsylvania, Philadelphia, Pennsylvania 19104; and ¢Department of Botany, University of Maryland, College Park, Maryland 20742 Abstract. Plasmodesmata or intercellular bridges that lyzed, and then fixed. As the cells retract from their connect plant ceils are cylindrical channels •40 nm in cell walls they leave behind the plasmodesmata still diameter. Running through the center of each is a inserted in the cell wall. They can break cleanly when dense rod, the desmotubule, that is connected to the the cell proper retracts or can pull away portions of endoplasmic reticulum of adjacent ceils. Fern, the plasma membrane of the cell with them. Where Onoclea sensibilis, gametophytes were cut in half and the desmotubule remains intact, the plasmodesma re- the cut surfaces exposed to the detergent, Triton X 100, tains its shape. These images and the results with de- then fixed. Although the plasma membrane limiting tergents and proteases indicate that the desmotubule the plasmodesma is solubilized partially or completely, provides a cytoskeletal element for each plasmodesma, the desmotubule remains intact. Alternatively, if the an element that not only stabilizes the whole struc- cut surface is exposed to papain, then fixed, the des- ture, but also limits its size and porosity. It is likely to motubule disappears, but the plasma membrane limit- be composed in large part of protein. Suggestions are ing the plasmodesmata remains intact albeit swollen made as to why this structure has been selected for in and irregular in profile. -

Apoplastic Route Cell Wall Symplastic Route Transmembrane Route Cytosol Key Plasmodesma Plasma Membrane Apoplast Symplast
CO O2 2 Light Sugar H2O O2 H2O and CO2 minerals © 2014 Pearson Education, Inc. 1 © 2014 Pearson Education, Inc. 2 Cell wall 24 32 42 29 40 16 Apoplastic route 11 19 21 27 34 8 3 6 Cytosol 14 13 Symplastic route 26 Shoot 1 5 apical 22 Transmembrane route meristem 9 Buds 18 10 4 31 2 17 23 7 12 Key 15 Plasmodesma 20 25 28 Plasma membrane Apoplast 1 mm Symplast © 2014 Pearson Education, Inc. 3 © 2014 Pearson Education, Inc. 4 CYTOPLASM EXTRACELLULAR + S H+ H FLUID H+ + H+ H + Hydrogen + H ion H H+ + S S H + H + Initial flaccid cell: + + H H H + ψP = 0 H+ H ψS = −0.7 H+ + S S S 0.4 M sucrose Proton H H+ ψ = −0.7 MPa Pure water: + solution: pump H ψP = 0 + ψP = 0 H /sucrose Sucrose Plasmolyzed ψS = 0 Turgid cell ψ = −0.9 (a) H+ and membrane potential cotransporter (neutral solute) cell at osmotic S ψ = 0 MPa at osmotic equilibrium ψ = −0.9 MPa equilibrium (b) H+ and cotransport of neutral solutes with its with its + + surroundings surroundings H − H 3 ψP = 0 ψP = 0.7 NO − + 3 H NO + ψS = −0.9 ψS = −0.7 + H+ K Potassium ion H ψ = −0.9 MPa ψ = 0 MPa + + H Nitrate K H+ K+ + − H+ K NO3 (a) Initial conditions: (b) Initial conditions: − + 3 NO − K cellular ψ > environmental ψ cellular ψ < environmental ψ 3 − NO + + NO3 K K H+ + − + H /NO3 + H cotransporter H Ion channel (c) H+ and cotransport of ions (d) Ion channels © 2014 Pearson Education, Inc. -

Seedless Plants 14.3: Seed Plants: Gymnosperms 14.4: Seed Plants: Angiosperms
Concepts of Biology Chapter 14 | Diversity of Plants 325 14 | DIVERSITY OF PLANTS Figure 14.1 Plants dominate the landscape and play an integral role in human societies. (a) Palm trees grow in tropical or subtropical climates; (b) wheat is a crop in most of the world; the flower of (c) the cotton plant produces fibers that are woven into fabric; the potent alkaloids of (d) the beautiful opium poppy have influenced human life both as a medicinal remedy and as a dangerously addictive drug. (credit a: modification of work by “3BoysInSanDiego”/Wikimedia Commons”; credit b: modification of work by Stephen Ausmus, USDA ARS; credit c: modification of work by David Nance, USDA ARS; credit d: modification of work by Jolly Janner) Chapter Outline 14.1: The Plant Kingdom 14.2: Seedless Plants 14.3: Seed Plants: Gymnosperms 14.4: Seed Plants: Angiosperms Introduction Plants play an integral role in all aspects of life on the planet, shaping the physical terrain, influencing the climate, and maintaining life as we know it. For millennia, human societies have depended on plants for nutrition and medicinal compounds, and for many industrial by-products, such as timber, paper, dyes, and textiles. Palms provide materials 326 Chapter 14 | Diversity of Plants including rattans, oils, and dates. Wheat is grown to feed both human and animal populations. The cotton boll flower is harvested and its fibers transformed into clothing or pulp for paper. The showy opium poppy is valued both as an ornamental flower and as a source of potent opiate compounds. Current evolutionary thought holds that all plants are monophyletic: that is, descendants of a single common ancestor.