Dcbld2/Esdn Is Essential for Proper Optic Tract Formation and Retinal Lamination Ryan Mears Joy University of Vermont
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

EGFR Phosphorylation of DCBLD2 Recruits TRAF6 and Stimulates AKT-Promoted Tumorigenesis
The Journal of Clinical Investigation RESEARCH ARTICLE EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis Haizhong Feng,1,2 Giselle Y. Lopez,3 Chung Kwon Kim,2 Angel Alvarez,2 Christopher G. Duncan,3 Ryo Nishikawa,4 Motoo Nagane,5 An-Jey A. Su,6 Philip E. Auron,6 Matthew L. Hedberg,7 Lin Wang,7 Jeffery J. Raizer,2 John A. Kessler,2 Andrew T. Parsa,8 Wei-Qiang Gao,1 Sung-Hak Kim,9 Mutsuko Minata,9 Ichiro Nakano,9 Jennifer R. Grandis,7 Roger E. McLendon,3 Darell D. Bigner,3 Hui-Kuan Lin,10 Frank B. Furnari,11 Webster K. Cavenee,11 Bo Hu,2 Hai Yan,3 and Shi-Yuan Cheng1,2 1State Key Laboratory of Oncogenes and Related Genes, Renji-Med X Clinical Stem Cell Research Center, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China. 2Department of Neurology and Northwestern Brain Tumor Institute, Center for Genetic Medicine, Robert H. Lurie Comprehensive Cancer Center, Northwestern University Feinberg School of Medicine, Chicago, Illinois, USA. 3Pediatric Brain Tumor Foundation Institute at Duke, The Preston Robert Tisch Brain Tumor Center, and Department of Pathology, Duke University Medical Center, Durham, North Carolina, USA. 4Department of Neuro-Oncology/Neurosurgery, International Medical Center, Saitama Medical University, Saitama, Japan. 5Department of Neurosurgery, Kyorin University, Tokyo, Japan. 6Department of Biological Sciences, Duquesne University, Pittsburgh, Pennsylvania, USA. 7Departments of Otolaryngology and Pharmacology and Chemical Biology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA. 8Department of Neurological Surgery and Northwestern Brain Tumor Institute, Robert H. Lurie Comprehensive Cancer Center, Northwestern University Feinberg School of Medicine, Chicago, Illinois, USA. -

Downloaded the “Top Edge” Version
bioRxiv preprint doi: https://doi.org/10.1101/855338; this version posted December 6, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Drosophila models of pathogenic copy-number variant genes show global and 2 non-neuronal defects during development 3 Short title: Non-neuronal defects of fly homologs of CNV genes 4 Tanzeen Yusuff1,4, Matthew Jensen1,4, Sneha Yennawar1,4, Lucilla Pizzo1, Siddharth 5 Karthikeyan1, Dagny J. Gould1, Avik Sarker1, Yurika Matsui1,2, Janani Iyer1, Zhi-Chun Lai1,2, 6 and Santhosh Girirajan1,3* 7 8 1. Department of Biochemistry and Molecular Biology, Pennsylvania State University, 9 University Park, PA 16802 10 2. Department of Biology, Pennsylvania State University, University Park, PA 16802 11 3. Department of Anthropology, Pennsylvania State University, University Park, PA 16802 12 4 contributed equally to work 13 14 *Correspondence: 15 Santhosh Girirajan, MBBS, PhD 16 205A Life Sciences Building 17 Pennsylvania State University 18 University Park, PA 16802 19 E-mail: [email protected] 20 Phone: 814-865-0674 21 1 bioRxiv preprint doi: https://doi.org/10.1101/855338; this version posted December 6, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 22 ABSTRACT 23 While rare pathogenic copy-number variants (CNVs) are associated with both neuronal and non- 24 neuronal phenotypes, functional studies evaluating these regions have focused on the molecular 25 basis of neuronal defects. -

EGFR Phosphorylation of DCBLD2 Recruits TRAF6 and Stimulates AKT-Promoted Tumorigenesis
EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis Haizhong Feng, … , Hai Yan, Shi-Yuan Cheng J Clin Invest. 2014;124(9):3741-3756. https://doi.org/10.1172/JCI73093. Research Article Oncology Aberrant activation of EGFR in human cancers promotes tumorigenesis through stimulation of AKT signaling. Here, we determined that the discoidina neuropilin-like membrane protein DCBLD2 is upregulated in clinical specimens of glioblastomas and head and neck cancers (HNCs) and is required for EGFR-stimulated tumorigenesis. In multiple cancer cell lines, EGFR activated phosphorylation of tyrosine 750 (Y750) of DCBLD2, which is located within a recently identified binding motif for TNF receptor-associated factor 6 (TRAF6). Consequently, phosphorylation of DCBLD2 Y750 recruited TRAF6, leading to increased TRAF6 E3 ubiquitin ligase activity and subsequent activation of AKT, thereby enhancing EGFR-driven tumorigenesis. Moreover, evaluation of patient samples of gliomas and HNCs revealed an association among EGFR activation, DCBLD2 phosphorylation, and poor prognoses. Together, our findings uncover a pathway in which DCBLD2 functions as a signal relay for oncogenic EGFR signaling to promote tumorigenesis and suggest DCBLD2 and TRAF6 as potential therapeutic targets for human cancers that are associated with EGFR activation. Find the latest version: https://jci.me/73093/pdf The Journal of Clinical Investigation RESEARCH ARTICLE EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis Haizhong Feng,1,2 Giselle Y. Lopez,3 Chung Kwon Kim,2 Angel Alvarez,2 Christopher G. Duncan,3 Ryo Nishikawa,4 Motoo Nagane,5 An-Jey A. Su,6 Philip E. Auron,6 Matthew L. Hedberg,7 Lin Wang,7 Jeffery J. -

Both LCCL-Domains of Human CRISPLD2 Have High Affinity for Lipid A
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Biochimie 97 (2014) 66e71 Contents lists available at ScienceDirect Biochimie journal homepage: www.elsevier.com/locate/biochi Research paper Both LCCL-domains of human CRISPLD2 have high affinity for lipid A Viktor Vásárhelyi, Mária Trexler, László Patthy* Institute of Enzymology, Research Centre for Natural Sciences, Hungarian Academy of Sciences, Budapest H-1518, PO Box 7, Hungary article info abstract Article history: The LCCL-domain is a recently defined protein module present in diverse extracellular multidomain Received 15 May 2013 proteins. Practically nothing is known about the molecular function of these domains; based on func- Accepted 20 September 2013 tional features of proteins harboring LCCL-domains it has been suggested that these domains might Available online 1 October 2013 function as lipopolysaccharide-binding domains. Here we show that the two LCCL-domains of human CRISPLD2 protein, a lipopolysaccharide-binding serum protein involved in defense against endotoxin Keywords: shock, have higher affinity for the lipid A, the toxic moiety of lipopolysaccharides than for ipopoly- CRISPLD2 protein saccharide. Our observation that the LCCL-domains of CRISPLD2 are specific for the toxic lipid A moiety of Endotoxin LCCL-domain the endotoxin suggests that it may block the interaction between endotoxins and the host endotoxin Lipid A receptors without interfering with the development of antibacterial immunity against the polysaccharide Lipopolysaccharide moiety of LPS. We suggest that the anti-inflammatory function of CRISPLD2 protein may account for its role in various pathological and developmental processes. Ó 2013 The Authors. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -
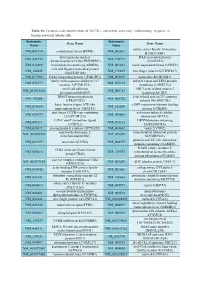
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

Rare and Low-Frequency Coding Variants Alter Human Adult Height
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Nature. Manuscript Author Author manuscript; Manuscript Author available in PMC 2017 August 01. Published in final edited form as: Nature. 2017 February 09; 542(7640): 186–190. doi:10.1038/nature21039. Rare and low-frequency coding variants alter human adult height A full list of authors and affiliations appears at the end of the article. Summary Height is a highly heritable, classic polygenic trait with ∼700 common associated variants identified so far through genome-wide association studies. Here, we report 83 height-associated coding variants with lower minor allele frequencies (range of 0.1-4.8%) and effects of up to 2 cm/ allele (e.g. in IHH, STC2, AR and CRISPLD2), >10 times the average effect of common variants. In functional follow-up studies, rare height-increasing alleles of STC2 (+1-2 cm/allele) compromised proteolytic inhibition of PAPP-A and increased cleavage of IGFBP-4 in vitro, resulting in higher bioavailability of insulin-like growth factors. These 83 height-associated variants overlap genes mutated in monogenic growth disorders and highlight new biological candidates (e.g. ADAMTS3, IL11RA, NOX4) and pathways (e.g. proteoglycan/ glycosaminoglycan synthesis) involved in growth. Our results demonstrate that sufficiently large sample sizes can uncover rare and low-frequency variants of moderate to large effect associated with polygenic human phenotypes, and that these variants implicate relevant genes and pathways. Reprints and permissions information is available at www.nature.com/reprints.Users may view, print, copy, and download text and data-mine the content in such documents, for the purposes of academic research, subject always to the full Conditions of use:http:// www.nature.com/authors/editorial_policies/license.html#terms Correspondence to: Joel N. -

A Homozygous Nonsense Mutation in DCBLD2 Is a Candidate Cause of Developmental Delay, Dysmorphic Features and Restrictive Cardiomyopathy Kheloud M
www.nature.com/scientificreports OPEN A homozygous nonsense mutation in DCBLD2 is a candidate cause of developmental delay, dysmorphic features and restrictive cardiomyopathy Kheloud M. Alhamoudi1, Tlili Barhoumi2, Hamad Al‑Eidi1, Abdulaziz Asiri3, Marwan Nashabat4, Manal Alaamery5, Masheal Alharbi1, Yazeid Alhaidan1, Brahim Tabarki6, Muhammad Umair1 & Majid Alfadhel1,4* DCBLD2 encodes discodin, CUB and LCCL domain‑containing protein 2, a type‑I transmembrane receptor that is involved in intracellular receptor signalling pathways and the regulation of cell growth. In this report, we describe a 5‑year‑old female who presented severe clinical features, including restrictive cardiomyopathy, developmental delay, spasticity and dysmorphic features. Trio‑ whole‑exome sequencing and segregation analysis were performed to identify the genetic cause of the disease within the family. A novel homozygous nonsense variant in the DCBLD2 gene (c.80G > A, p.W27*) was identifed as the most likely cause of the patient’s phenotype. This nonsense variant falls in the extracellular N‑terminus of DCBLD2 and thus might afect proper protein function of the transmembrane receptor. A number of in vitro investigations were performed on the proband’s skin fbroblasts compared to normal fbroblasts, which allowed a comprehensive assessment resulting in the functional characterization of the identifed DCBLD2 nonsense variant in diferent cellular processes. Our data propose a signifcant association between the identifed variant and the observed reduction in cell proliferation, -

Analysis of the Plasmodium Falciparum Proteome by High-Accuracy Mass Organism
CORE Metadata, citation and similar papers at core.ac.uk Provided by University of Southern Denmark Research Output letters to nature 12. Lasonder, E. et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass organism. Here we describe a large-scale, high-accuracy (average spectrometry. Nature 419, 537–542 (2002). 13. Ewing, B. & Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. deviation less than 0.02 Da at 1,000 Da) mass spectrometric 1–3 Genome Res. 8, 186–194 (1998). proteome analysis of selected stages of the human malaria 14. Oefner, P. J. et al. Efficient random subcloning of DNA sheared in a recirculating point-sink flow parasite Plasmodium falciparum. The analysis revealed 1,289 system. Nucleic Acids Res. 24, 3879–3886 (1996). proteins of which 714 proteins were identified in asexual blood 15. Ewing, B., Hillier, L., Wendl, M. C. & Green, P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185 (1998). stages, 931 in gametocytes and 645 in gametes. The last two 16. Gordon, D., Abajian, C. & Green, P. Consed: a graphical tool for sequence finishing. Genome Res. 8, groups provide insights into the biology of the sexual stages of 195–202 (1998). the parasite, and include conserved, stage-specific, secreted and 17. Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. membrane-associated proteins. A subset of these proteins con- J. Mol. Biol. 215, 403–410 (1990). 18. Apweiler, R. et al. The InterPro database, an integrated documentation resource for protein families, tain domains that indicate a role in cell–cell interactions, and domains and functional sites. -

Sun, B. B., Maranville, J. C., Peters, J. E., Stacey, D., Staley, J. R., Blackshaw, J., Burgess, S., Jiang, T., Paige, E., Suren
Sun, B. B., Maranville, J. C., Peters, J. E., Stacey, D., Staley, J. R., Blackshaw, J., Burgess, S., Jiang, T., Paige, E., Surendran, P., Oliver- Williams, C., Kamat, M. A., Prins, B. P., Wilcox, S. K., Zimmerman, E. S., Chi, A., Bansal, N., Spain, S. L., Wood, A. M., ... Butterworth, A. S. (2018). Genomic atlas of the human plasma proteome. Nature, 558(7708), 73-79. https://doi.org/10.1038/s41586-018-0175-2 Peer reviewed version Link to published version (if available): 10.1038/s41586-018-0175-2 Link to publication record in Explore Bristol Research PDF-document This is the author accepted manuscript (AAM). The final published version (version of record) is available online via Nature Publishing Group at https://www.nature.com/articles/s41586-018-0175-2 . Please refer to any applicable terms of use of the publisher. University of Bristol - Explore Bristol Research General rights This document is made available in accordance with publisher policies. Please cite only the published version using the reference above. Full terms of use are available: http://www.bristol.ac.uk/red/research-policy/pure/user-guides/ebr-terms/ Genomic atlas of the human plasma proteome Benjamin B. Sun1*, Joseph C. Maranville2*, James E. Peters1,3*, David Stacey1, James R. Staley1, James Blackshaw1, Stephen Burgess1,4, Tao Jiang1, Ellie Paige1,5, Praveen Surendran1, Clare Oliver-Williams1,6, Mihir A. Kamat1, Bram P. Prins1, Sheri K. Wilcox7, Erik S. Zimmerman7, An Chi2, Narinder Bansal1,8, Sarah L. Spain9, Angela M. Wood1, Nicholas W. Morrell10, John R. Bradley11, Nebojsa Janjic7, David J. Roberts12,13, Willem H. -

Mouse Dcbld2 Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Dcbld2 Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Dcbld2 conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Dcbld2 gene (NCBI Reference Sequence: NM_028523 ; Ensembl: ENSMUSG00000035107 ) is located on Mouse chromosome 16. 16 exons are identified, with the ATG start codon in exon 1 and the TGA stop codon in exon 16 (Transcript: ENSMUST00000046663). Exon 5~7 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Dcbld2 gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP23-86O4 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mice homozygous for a knock-out allele exhibit reduced postnatal angiogenesis and impaired recovery from femoral artery ligation with impaired blood flow and decreased capillary density. Exon 5 starts from about 26.66% of the coding region. The knockout of Exon 5~7 will result in frameshift of the gene. The size of intron 4 for 5'-loxP site insertion: 3059 bp, and the size of intron 7 for 3'-loxP site insertion: 884 bp. The size of effective cKO region: ~1888 bp. The cKO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 5 6 7 8 9 16 Targeting vector Targeted allele Constitutive KO allele (After Cre recombination) Legends Exon of mouse Dcbld2 Homology arm cKO region loxP site Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Cleaved Cochlin Sequesters Pseudomonas Aeruginosa and Activates Innate Immunity in the Inner Ear
Article Cleaved Cochlin Sequesters Pseudomonas aeruginosa and Activates Innate Immunity in the Inner Ear Graphical Abstract Authors Jinsei Jung, Jee Eun Yoo, Young Ho Choe, ..., Joo-Heon Yoon, Young-Min Hyun, Jae Young Choi Correspondence [email protected] (Y.-M.H.), [email protected] (J.Y.C.) In Brief Jung et al. show that the cleaved cochlin LCCL domain enhances innate immune responses in the inner ear by aggregating infiltrated bacteria and recruiting innate immune cells. This spatiotemporally protective function of LCCL protects hearing function in the organ of Corti against effects of bacterial invasion in the inner ear. Highlights d Cleaved inner ear cochlin LCCL secretes to perilymph space post-bacterial infection d LCCL induces bacterial aggregation in the scala tympani d Spatiotemporal innate immune response by LCCL protects the sensory organ of Corti À À d LCCL rescues Coch / mouse post-Pseudomonas inner ear infection hearing loss Jung et al., 2019, Cell Host & Microbe 25, 1–13 April 10, 2019 ª 2019 Elsevier Inc. https://doi.org/10.1016/j.chom.2019.02.001 Please cite this article in press as: Jung et al., Cleaved Cochlin Sequesters Pseudomonas aeruginosa and Activates Innate Immunity in the Inner Ear, Cell Host & Microbe (2019), https://doi.org/10.1016/j.chom.2019.02.001 Cell Host & Microbe Article Cleaved Cochlin Sequesters Pseudomonas aeruginosa and Activates Innate Immunity in the Inner Ear Jinsei Jung,1,2,7 Jee Eun Yoo,1,2,7 Young Ho Choe,3,4 Sang Chul Park,1 Hyun Jae Lee,1,2 Hack June Lee,1,2 Byunghwa Noh,1,2 Sung Huhn Kim,1,2