Domain Shuffling and the Evolution of Vertebrates
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Both LCCL-Domains of Human CRISPLD2 Have High Affinity for Lipid A
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Biochimie 97 (2014) 66e71 Contents lists available at ScienceDirect Biochimie journal homepage: www.elsevier.com/locate/biochi Research paper Both LCCL-domains of human CRISPLD2 have high affinity for lipid A Viktor Vásárhelyi, Mária Trexler, László Patthy* Institute of Enzymology, Research Centre for Natural Sciences, Hungarian Academy of Sciences, Budapest H-1518, PO Box 7, Hungary article info abstract Article history: The LCCL-domain is a recently defined protein module present in diverse extracellular multidomain Received 15 May 2013 proteins. Practically nothing is known about the molecular function of these domains; based on func- Accepted 20 September 2013 tional features of proteins harboring LCCL-domains it has been suggested that these domains might Available online 1 October 2013 function as lipopolysaccharide-binding domains. Here we show that the two LCCL-domains of human CRISPLD2 protein, a lipopolysaccharide-binding serum protein involved in defense against endotoxin Keywords: shock, have higher affinity for the lipid A, the toxic moiety of lipopolysaccharides than for ipopoly- CRISPLD2 protein saccharide. Our observation that the LCCL-domains of CRISPLD2 are specific for the toxic lipid A moiety of Endotoxin LCCL-domain the endotoxin suggests that it may block the interaction between endotoxins and the host endotoxin Lipid A receptors without interfering with the development of antibacterial immunity against the polysaccharide Lipopolysaccharide moiety of LPS. We suggest that the anti-inflammatory function of CRISPLD2 protein may account for its role in various pathological and developmental processes. Ó 2013 The Authors. -
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
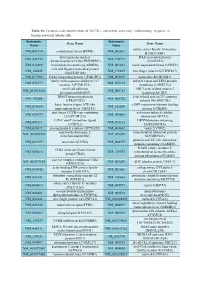
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

Analysis of the Plasmodium Falciparum Proteome by High-Accuracy Mass Organism
CORE Metadata, citation and similar papers at core.ac.uk Provided by University of Southern Denmark Research Output letters to nature 12. Lasonder, E. et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass organism. Here we describe a large-scale, high-accuracy (average spectrometry. Nature 419, 537–542 (2002). 13. Ewing, B. & Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. deviation less than 0.02 Da at 1,000 Da) mass spectrometric 1–3 Genome Res. 8, 186–194 (1998). proteome analysis of selected stages of the human malaria 14. Oefner, P. J. et al. Efficient random subcloning of DNA sheared in a recirculating point-sink flow parasite Plasmodium falciparum. The analysis revealed 1,289 system. Nucleic Acids Res. 24, 3879–3886 (1996). proteins of which 714 proteins were identified in asexual blood 15. Ewing, B., Hillier, L., Wendl, M. C. & Green, P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185 (1998). stages, 931 in gametocytes and 645 in gametes. The last two 16. Gordon, D., Abajian, C. & Green, P. Consed: a graphical tool for sequence finishing. Genome Res. 8, groups provide insights into the biology of the sexual stages of 195–202 (1998). the parasite, and include conserved, stage-specific, secreted and 17. Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. membrane-associated proteins. A subset of these proteins con- J. Mol. Biol. 215, 403–410 (1990). 18. Apweiler, R. et al. The InterPro database, an integrated documentation resource for protein families, tain domains that indicate a role in cell–cell interactions, and domains and functional sites. -

Identification of a Heterozygous ACAN Mutation in a 15-Year-Old Boy With
Case report https://doi.org/10.6065/apem.1938198.099 Ann Pediatr Endocrinol Metab 2020;25:272-276 Identification of a heterozygous ACAN mutation in a 15-year-old boy with short stature who presented with advanced bone age: a case report and review of the literature Tae Youp Kim, MD1, Longitudinal bone growth is primarily mediated by the growth plate, which is Kyung Mi Jang, MD, PhD1, a specialized cartilaginous structure. Aggrecan, encoded by ACAN, is a primary Chang Won Keum, PhD2, proteoglycan component of the extracellular matrix in both the growth plate and Seung Hwan Oh, MD, PhD3, articular cartilage. Aggrecanopathies have emerged as a phenotype of genetic Woo Yeong Chung, MD, PhD4 skeletal disease in humans. A heterozygous ACAN mutation causes short stature, premature growth cessation, and accelerated bone age maturation. We report 1Department of Pediatrics, Yeungnam the case of a 15-year-old boy with familial short stature, with height of 149 cm University Hospital, Yeungnam Univer- (Korean standard deviation score [SDS] of -3.6) and weight of 50.5 kg (-1.48 SDS). sity College of Medicine, Daegu, Korea He presented with mild midfacial hypoplasia, frontal bossing, a broad chest, and 2Rare Genetic Disease Research Center, a short neck. The father's and mother's heights were 150 cm (-4.8 SDS) and 153 3Billion INC, Seoul, Korea cm (-1.69 SDS), respectively. The patient's bone age was 2–3 years more advanced 3 Departments of Laboratory Medicine than his chronological age, and no endocrine abnormalities were detected. Whole- 4 and Pediatrics, Inje University Busan exome sequencing followed by Sanger sequencing revealed a heterozygous ACAN Paik Hospital, Inje University College of mutation, c.512C>T (p.Ala171Val), in both the proband and his father. -

Structural Characterisation of Aggrecan in Cartilaginous Tissues and Tissue Engineered Constructs a Thesis Submitted to the Univ
Structural Characterisation of Aggrecan in Cartilaginous Tissues and Tissue Engineered Constructs A thesis submitted to the University of Manchester for the degree of Doctor of Philosophy in the Faculty of Biology, Medicine and Health 2017 Russell J Craddock The School of Biological Sciences Division of Cell Matrix Biology and Regenerative Medicine List of Contents List of Contents ................................................................................................................. 2 List of Figures ................................................................................................................... 6 List of Tables ..................................................................................................................... 8 List of Equations ............................................................................................................... 8 List of Conference Presentations and Publications ....................................................... 8 List of Abbreviations ........................................................................................................ 9 Abstract ........................................................................................................................... 10 Declaration ...................................................................................................................... 11 Copyright Statement ...................................................................................................... 11 Acknowledgements ........................................................................................................ -

Binding of Hyaluronan and Its Neutral Analog by TSG-6 Link Domain
Binding of hyaluronan and its neutral analog by TSG-6 Link domain Roman Witasek 1, Eva Kutálková 1, Josef Hrnčiřík 1 and Marek Ingr 1,2 1Tomas Bata University in Zlín, Faculty of Technology, Department of Physics and Materials Engineering, Nám. T.G. Masaryka 5555, 76001 Zlín, Czech Republic 2Charles University, Faculty of Science, Department of Biochemistry, Hlavova 8/2030, 12843 Praha 2, Czech Republic 2nd Users’ Conference of Zlín IT4Innovations −> −> Hyaluronic acid (hyaluronan) – HA [4)-β-D-Glc pA-(1 3)-β-D-Glc pNAc-(1 ]n 1→3 1→4 1→3 1→4 1→4 • Essential component of proteoglycan - the main constituent of the extracellular matrix of connective tissues • Signaling molecule involved in carcinogenesis, inflammation and wound healing • Biological activity depends on the chain length (from MDa to short oligosaccharides) http://medinfo.ufl.edu/pa/chuck/summer/handouts/images/gag.jpg Hyaladherins – hyaluronan binding proteins TSG-6 Containing Link domain Consists of Link and CUB domain • Proteoglycan forming Link binds hyaluronan and other GAGs Aggrecan positively charged Brevican • CUB Neurocan not well known function Versican negatively charged Membrane receptors CUB CD44 – the main transmembrane receptor Link LYVE-1 – lymphatic endothelial cells Soluble receptor TSG-6 Without Link domain RHAMM – membrane receptor, partially disordered Inter-α-inhibitor Interactions of TSG-6 with glycosaminoglycans (GAGs) Several interactions of GAG oligosaccharides were identified by NMR experiments Computational details: Hyaluronan 1: K11, Y12, H45, V57, Y59, P60, I61, K63, F70, I76, Y78, R81, W88 • MD simulations carried out 2 st in GROMACS v. 5.1.1 Heparin : K34, K54, R56 + 1 mode: K20, K41, R84 ; • nd CHARMM 36 force field 2 mode: K72 • TIP3P model of water Chondroitin sulfate 3: 1st mode similar to HA ; • Water box 8 ×8×8 nm 2nd mode: K20, K34, K41, K54 • HA (GlcHA) oligosaccharides of 12 Questions: monosacharide units • TSG-6 Link domain – PDB code: 2N40 1. -

Tnfα-Stimulated Gene-6 (TSG6) Activates Macrophage PNAS PLUS Phenotype Transition to Prevent Inflammatory Lung Injury
TNFα-stimulated gene-6 (TSG6) activates macrophage PNAS PLUS phenotype transition to prevent inflammatory lung injury Manish Mittala, Chinnaswamy Tiruppathia, Saroj Nepala, You-Yang Zhaoa, Dagmara Grzycha, Dheeraj Sonia, Darwin J. Prockopb,1, and Asrar B. Malika,1 aDepartment of Pharmacology and Center for Lung and Vascular Biology, University of Illinois College of Medicine, Chicago, IL 60612 and bInstitute for Regenerative Medicine, Texas A&M University Health Science Center and College of Medicine, Bryan, TX 77807 Contributed by Darwin J. Prockop, November 3, 2016 (sent for review September 7, 2016; reviewed by Arshad Rehman and Sarah Yuan) TNFα-stimulated gene-6 (TSG6), a 30-kDa protein generated by acti- macrophages and other inflammatory cells sequestered in lungs (15). vated macrophages, modulates inflammation; however, its mechanism Macrophages in ALI are activated through the sensing of pathogen- of action and role in the activation of macrophages are not fully un- associated molecular patterns (PAMPs) and damage-associated derstood. Here we observed markedly augmented LPS-induced inflam- molecular patterns (DAMPs) via Toll-like receptors (TLRs). On −/− matory lung injury and mortality in TSG6 mice compared with WT binding of LPS to TLR4, macrophages produce proinflammatory +/+ (TSG6 ) mice. Treatment of mice with intratracheal instillation of cytokines, such as IFNγ,TNFα,andIL-1β,whichsignalthere- TSG6 prevented LPS-induced lung injury and neutrophil sequestration, cruitmentofneutrophilsandlymphocytes at the site of infection -

Ligand Binding and Signaling of HARE/Stabilin-2
biomolecules Review Ligand Binding and Signaling of HARE/Stabilin-2 Edward N. Harris * and Fatima Cabral Department of Biochemistry, University of Nebraska, Lincoln, NE 68588, USA * Correspondence: [email protected]; Tel.: +1-(402)-472-7468 Received: 18 June 2019; Accepted: 7 July 2019; Published: 11 July 2019 Abstract: The Stabilin receptors are a two-member family in the type H class of scavenger receptors. These dynamic receptors bind and internalize multiple ligands from the cell surface for the purpose of clearing extracellular material including some synthetic drugs and for sensing the external environment of the cell. Stabilin-1 was the first receptor to be cloned, though the biological activity of Hyaluronic Acid Receptor for Endocytosis (HARE)/Stabilin-2 was observed about 10 years prior to the cloning of Stabilin-1. Stabilin-1 has a more diverse expression profile among the tissues than HARE/Stabilin-2. This review will focus on HARE/Stabilin-2 and its interactions with hyaluronan, heparin, and phosphorothioate antisense oligonucleotides and what is known about how this receptor participates in signaling upon ligand binding. Keywords: stabilin; HARE; receptor-mediated endocytosis; scavenger receptor; hyaluronan 1. Initial Discovery The discovery of the cloned Stabilin receptors began with Stabilin-1 in the early 1990s. A monoclonal antibody raised against whole human spleen homogenate produced a monoclonal antibody (MS-1) against an antigen that was initially identified as a large protein that is preferentially expressed in the sinusoidal endothelia of spleen, lymph node, liver, and adrenal cortex [1]. The identified MS-1 antigen is an inducible protein not found in normal skin lesions, but rather in diseased skin and some cancers such as psoriasis, melanoma, and T-cell lymphomas. -

Cleaved Cochlin Sequesters Pseudomonas Aeruginosa and Activates Innate Immunity in the Inner Ear
Article Cleaved Cochlin Sequesters Pseudomonas aeruginosa and Activates Innate Immunity in the Inner Ear Graphical Abstract Authors Jinsei Jung, Jee Eun Yoo, Young Ho Choe, ..., Joo-Heon Yoon, Young-Min Hyun, Jae Young Choi Correspondence [email protected] (Y.-M.H.), [email protected] (J.Y.C.) In Brief Jung et al. show that the cleaved cochlin LCCL domain enhances innate immune responses in the inner ear by aggregating infiltrated bacteria and recruiting innate immune cells. This spatiotemporally protective function of LCCL protects hearing function in the organ of Corti against effects of bacterial invasion in the inner ear. Highlights d Cleaved inner ear cochlin LCCL secretes to perilymph space post-bacterial infection d LCCL induces bacterial aggregation in the scala tympani d Spatiotemporal innate immune response by LCCL protects the sensory organ of Corti À À d LCCL rescues Coch / mouse post-Pseudomonas inner ear infection hearing loss Jung et al., 2019, Cell Host & Microbe 25, 1–13 April 10, 2019 ª 2019 Elsevier Inc. https://doi.org/10.1016/j.chom.2019.02.001 Please cite this article in press as: Jung et al., Cleaved Cochlin Sequesters Pseudomonas aeruginosa and Activates Innate Immunity in the Inner Ear, Cell Host & Microbe (2019), https://doi.org/10.1016/j.chom.2019.02.001 Cell Host & Microbe Article Cleaved Cochlin Sequesters Pseudomonas aeruginosa and Activates Innate Immunity in the Inner Ear Jinsei Jung,1,2,7 Jee Eun Yoo,1,2,7 Young Ho Choe,3,4 Sang Chul Park,1 Hyun Jae Lee,1,2 Hack June Lee,1,2 Byunghwa Noh,1,2 Sung Huhn Kim,1,2 -

Subcellular Localisation, Secretion, and Post-Translational Processing Of
479 ORIGINAL ARTICLE J Med Genet: first published as 10.1136/jmg.40.7.479 on 1 July 2003. Downloaded from Subcellular localisation, secretion, and post-translational processing of normal cochlin, and of mutants causing the sensorineural deafness and vestibular disorder, DFNA9 N G Robertson, S A Hamaker, V Patriub, J C Aster, C C Morton ............................................................................................................................. J Med Genet 2003;40:479–486 Five missense mutations in the FCH/LCCL domain of the COCH gene, encoding the protein cochlin, are pathogenic for the autosomal dominant hearing loss and vestibular dysfunction disorder, DFNA9. To date, the function of cochlin and the mechanism of pathogenesis of the mutations are unknown. We have used the biological system of transient transfections of the entire protein coding region of COCH into several mammalian cell lines, to investigate various functional properties of cochlin. By western blot analysis of lysates prepared from transfected cells, we show that cochlin is a secreted protein. Immuno- cytochemistry shows concentrated localisation of cochlin in perinuclear structures consistent with the Golgi apparatus and endoplasmic reticulum, showing intracellular passage through these secretory compartments. We detected that cochlin is proteolytically cleaved between the FCH/LCCL domain and the downstream vWFA domains, resulting in a smaller cochlin isoform of ∼50 kDa. Interestingly, this isoform lacks the entire mutation bearing FCH/LCCL domain. We have also shown that cochlin is See end of article for N-glycosylated in its mature secreted form. Previous studies of the FCH/LCCL domain alone, expressed authors’ affiliations in bacteria, have demonstrated that three of four DFNA9 mutations cause misfolding of this domain. -

Does Otovestibular Loss in the Autosomal Dominant Disorder DFNA9 Have an Impact of on Cognition? a Systematic Review
SYSTEMATIC REVIEW published: 09 January 2018 doi: 10.3389/fnins.2017.00735 Does Otovestibular Loss in the Autosomal Dominant Disorder DFNA9 Have an Impact of on Cognition? A Systematic Review Jonas De Belder 1†, Stijn Matthysen 1†, Annes J. Claes 1, 2, Griet Mertens 1, 2, Paul Van de Heyning 1, 2 and Vincent Van Rompaey 1, 2* 1 Faculty of Medicine and Health Sciences, University of Antwerp, Antwerp, Belgium, 2 Department of Otorhinolaryngology and Head and Neck Surgery, Antwerp University Hospital, Edegem, Belgium Edited by: Background and Purpose: Cognitive impairment has been observed in patients with Etienne De Villers-Sidani, bilateral vestibular loss (BVL) and in patients with sensorineural hearing loss (SNHL). McGill University, Canada DFNA9 is an autosomal dominant disorder that causes a combination of both sensory Reviewed by: deficits by the 3rd to 5th decade. We therefore hypothesize a combined detrimental effect Martha M. Shiell, Maastricht University, Netherlands on cognition. The aim of this systematic review was to identify studies related to DFNA9 Ronald Pennings, in general and its relationship with cognitive impairment more specifically. Radboud University Nijmegen Medical Center, Netherlands Materials and Methods: Several databases including Medline, Cochrane Database *Correspondence: of Systematic Reviews, Cochrane Central Register of Controlled Trials, ISI Web of Vincent Van Rompaey Knowledge, and Web of Science were searched to accumulate information about [email protected] DFNA9-mutations, including phenotype, genotype, pathophysiology, quality of life (QOL), †These authors have contributed equally to this work. and imaging in general and cognitive function more specifically. A qualitative analysis was performed on the 55 articles that qualified. -

CXCL8 Direct Interaction with the Chemokine TSG-6 Inhibits
TSG-6 Inhibits Neutrophil Migration via Direct Interaction with the Chemokine CXCL8 This information is current as Douglas P. Dyer, Jennifer M. Thomson, Aurelie Hermant, of September 23, 2021. Thomas A. Jowitt, Tracy M. Handel, Amanda E. I. Proudfoot, Anthony J. Day and Caroline M. Milner J Immunol 2014; 192:2177-2185; Prepublished online 5 February 2014; doi: 10.4049/jimmunol.1300194 Downloaded from http://www.jimmunol.org/content/192/5/2177 Supplementary http://www.jimmunol.org/content/suppl/2014/02/05/jimmunol.130019 Material 4.DCSupplemental http://www.jimmunol.org/ References This article cites 56 articles, 27 of which you can access for free at: http://www.jimmunol.org/content/192/5/2177.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on September 23, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2014 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology TSG-6 Inhibits Neutrophil Migration via Direct Interaction with the Chemokine CXCL8 Douglas P.