Supp. Material-Table1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reconstitution of Long and Short Patch Mismatch Repair Reactions Using Saccharomyces Cerevisiae Proteins
Reconstitution of long and short patch mismatch repair reactions using Saccharomyces cerevisiae proteins Nikki Bowena, Catherine E. Smitha, Anjana Srivatsana, Smaranda Willcoxb,c, Jack D. Griffithb,c, and Richard D. Kolodnera,d,e,f,g,1 aLudwig Institute for Cancer Research, Departments of dMedicine and eCellular and Molecular Medicine, fMoores-University of California, San Diego Cancer Center, and gInstitute of Genomic Medicine, University of California San Diego School of Medicine, La Jolla, CA 92093; and bLineberger Cancer Center and cDepartment of Microbiology and Immunology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27514 Contributed by Richard D. Kolodner, October 8, 2013 (sent for review September 9, 2013) A problem in understanding eukaryotic DNA mismatch repair In eukaryotic MMR, mispairs are bound by MutS homolog 2 (MMR) mechanisms is linking insights into MMR mechanisms from (Msh2)–MutS homolog 6 (Msh6) and Msh2–MutS homolog 3 genetics and cell-biology studies with those from biochemical (Msh3), two partially redundant complexes of MutS-related pro- studies of MMR proteins and reconstituted MMR reactions. This teins (3, 4, 18, 19). These complexes recruit a MutL-related type of analysis has proven difficult because reconstitution ap- complex, called MutL homoloh 1 (Mlh1)–postmeiotic segrega- proaches have been most successful for human MMR whereas tion 1 (Pms1) in S. cerevisiae and Mlh1–postmeiotic segregation – – analysis of MMR in vivo has been most advanced in the yeast 2 (Pms2) in human and mouse (3, 4, 20 23). The Mlh1 Pms1/ Saccharomyces cerevisiae. Here, we describe the reconstitution of Pms2 complex has an endonuclease activity suggested to play MMR reactions using purified S. -

Paul Modrich Howard Hughes Medical Institute and Department of Biochemistry, Duke University Medical Center, Durham, North Carolina, USA
Mechanisms in E. coli and Human Mismatch Repair Nobel Lecture, December 8, 2015 by Paul Modrich Howard Hughes Medical Institute and Department of Biochemistry, Duke University Medical Center, Durham, North Carolina, USA. he idea that mismatched base pairs occur in cells and that such lesions trig- T ger their own repair was suggested 50 years ago by Robin Holliday in the context of genetic recombination [1]. Breakage and rejoining of DNA helices was known to occur during this process [2], with precision of rejoining attributed to formation of a heteroduplex joint, a region of helix where the two strands are derived from the diferent recombining partners. Holliday pointed out that if this heteroduplex region should span a genetic diference between the two DNAs, then it will contain one or more mismatched base pairs. He invoked processing of such mismatches to explain the recombination-associated phenomenon of gene conversion [1], noting that “If there are enzymes which can repair points of damage in DNA, it would seem possible that the same enzymes could recognize the abnormality of base pairing, and by exchange reactions rectify this.” Direct evidence that mismatches provoke a repair reaction was provided by bacterial transformation experiments [3–5], and our interest in this efect was prompted by the Escherichia coli (E. coli) work done in Matt Meselson’s lab at Harvard. Using artifcially constructed heteroduplex DNAs containing multiple mismatched base pairs, Wagner and Meselson [6] demonstrated that mismatches elicit a repair reaction upon introduction into the E. coli cell. Tey also showed that closely spaced mismatches, mismatches separated by a 1000 base pairs or so, are usually repaired on the same DNA strand. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

Molecular Basis for the Distinct Cellular Functions of the Lsm1-7 and Lsm2-8 Complexes
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.22.055376; this version posted April 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes Eric J. Montemayor1,2, Johanna M. Virta1, Samuel M. Hayes1, Yuichiro Nomura1, David A. Brow2, Samuel E. Butcher1 1Department of Biochemistry, University of Wisconsin-Madison, Madison, WI, USA. 2Department of Biomolecular Chemistry, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA. Correspondence should be addressed to E.J.M. ([email protected]) and S.E.B. ([email protected]). Abstract Eukaryotes possess eight highly conserved Lsm (like Sm) proteins that assemble into circular, heteroheptameric complexes, bind RNA, and direct a diverse range of biological processes. Among the many essential functions of Lsm proteins, the cytoplasmic Lsm1-7 complex initiates mRNA decay, while the nuclear Lsm2-8 complex acts as a chaperone for U6 spliceosomal RNA. It has been unclear how these complexes perform their distinct functions while differing by only one out of seven subunits. Here, we elucidate the molecular basis for Lsm-RNA recognition and present four high-resolution structures of Lsm complexes bound to RNAs. The structures of Lsm2-8 bound to RNA identify the unique 2′,3′ cyclic phosphate end of U6 as a prime determinant of specificity. In contrast, the Lsm1-7 complex strongly discriminates against cyclic phosphates and tightly binds to oligouridylate tracts with terminal purines. -

Phosphate Steering by Flap Endonuclease 1 Promotes 50-flap Specificity and Incision to Prevent Genome Instability
ARTICLE Received 18 Jan 2017 | Accepted 5 May 2017 | Published 27 Jun 2017 DOI: 10.1038/ncomms15855 OPEN Phosphate steering by Flap Endonuclease 1 promotes 50-flap specificity and incision to prevent genome instability Susan E. Tsutakawa1,*, Mark J. Thompson2,*, Andrew S. Arvai3,*, Alexander J. Neil4,*, Steven J. Shaw2, Sana I. Algasaier2, Jane C. Kim4, L. David Finger2, Emma Jardine2, Victoria J.B. Gotham2, Altaf H. Sarker5, Mai Z. Her1, Fahad Rashid6, Samir M. Hamdan6, Sergei M. Mirkin4, Jane A. Grasby2 & John A. Tainer1,7 DNA replication and repair enzyme Flap Endonuclease 1 (FEN1) is vital for genome integrity, and FEN1 mutations arise in multiple cancers. FEN1 precisely cleaves single-stranded (ss) 50-flaps one nucleotide into duplex (ds) DNA. Yet, how FEN1 selects for but does not incise the ss 50-flap was enigmatic. Here we combine crystallographic, biochemical and genetic analyses to show that two dsDNA binding sites set the 50polarity and to reveal unexpected control of the DNA phosphodiester backbone by electrostatic interactions. Via ‘phosphate steering’, basic residues energetically steer an inverted ss 50-flap through a gateway over FEN1’s active site and shift dsDNA for catalysis. Mutations of these residues cause an 18,000-fold reduction in catalytic rate in vitro and large-scale trinucleotide (GAA)n repeat expansions in vivo, implying failed phosphate-steering promotes an unanticipated lagging-strand template-switch mechanism during replication. Thus, phosphate steering is an unappreciated FEN1 function that enforces 50-flap specificity and catalysis, preventing genomic instability. 1 Molecular Biophysics and Integrated Bioimaging, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA. -

Identification of Molecular Targets in Head and Neck Squamous Cell Carcinomas Based on Genome-Wide Gene Expression Profiling
1489-1497 7/11/07 18:41 Page 1489 ONCOLOGY REPORTS 18: 1489-1497, 2007 Identification of molecular targets in head and neck squamous cell carcinomas based on genome-wide gene expression profiling SATOYA SHIMIZU1,2, NAOHIKO SEKI2, TAKASHI SUGIMOTO2, SHIGETOSHI HORIGUCHI1, HIDEKI TANZAWA3, TOYOYUKI HANAZAWA1 and YOSHITAKA OKAMOTO1 Departments of 1Otorhinolaryngology, 2Functional Genomics and 3Clinical Molecular Biology, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan Received May 21, 2007; Accepted June 28, 2007 Abstract. DNA amplifications activate oncogenes and are patients and metastases develop in 15-25% of patients (1). hallmarks of nearly all advanced cancers including head and Many factors, such as TNM stage, pathological grade and neck squamous cell carcinoma (HNSCC). Some oncogenes tumor site, influence the prognosis of HNSCC but are not show both DNA copy number gain and mRNA overexpression. sufficient to predict outcome. In addition, treatment often Chromosomal comparative genomic hybridization and oligo- results in impairment of functions such as speech and nucleotide microarrays were used to examine 8 HNSCC cell swallowing, cosmetic disfiguration and mental pain. These lines and a plot of gene expression levels relative to their inflictions significantly erode quality of life. To overcome this position on the chromosome was produced. Three highly situation, there is a need to find novel biomarkers that classify up-regulated genes, NT5C3, ANLN and INHBA, were patients into prognostic groups, to aid identification of high- identified on chromosome 7p14. These genes were subjected risk patients who may benefit from different treatments. to quantitative real-time RT-PCR on cDNA and genomic Comparative genomic hybridization (CGH) has facilitated DNA derived from 8 HNSCC cell lines. -
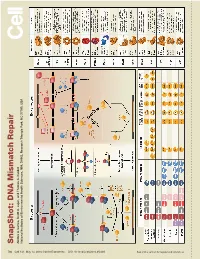
S Na P S H O T: D N a Mism a Tc H R E P a Ir
SnapShot: Repair DNA Mismatch Scott A. Lujan, and Thomas Kunkel A. Larrea, Andres Park, NC 27709, USA Triangle Health Sciences, NIH, DHHS, Research National Institutes of Environmental 730 Cell 141, May 14, 2010 ©2010 Elsevier Inc. DOI 10.1016/j.cell.2010.05.002 See online version for legend and references. SnapShot: DNA Mismatch Repair Andres A. Larrea, Scott A. Lujan, and Thomas A. Kunkel National Institutes of Environmental Health Sciences, NIH, DHHS, Research Triangle Park, NC 27709, USA Mismatch Repair in Bacteria and Eukaryotes Mismatch repair in the bacterium Escherichia coli is initiated when a homodimer of MutS binds as an asymmetric clamp to DNA containing a variety of base-base and insertion-deletion mismatches. The MutL homodimer then couples MutS recognition to the signal that distinguishes between the template and nascent DNA strands. In E. coli, the lack of adenine methylation, catalyzed by the DNA adenine methyltransferase (Dam) in newly synthesized GATC sequences, allows E. coli MutH to cleave the nascent strand. The resulting nick is used for mismatch removal involving the UvrD helicase, single-strand DNA-binding protein (SSB), and excision by single-stranded DNA exonucleases from either direction, depending upon the polarity of the nick relative to the mismatch. DNA polymerase III correctly resynthesizes DNA and ligase completes repair. In bacteria lacking Dam/MutH, as in eukaryotes, the signal for strand discrimination is uncertain but may be the DNA ends associated with replication forks. In these bacteria, MutL harbors a nick-dependent endonuclease that creates a nick that can be used for mismatch excision. Eukaryotic mismatch repair is similar, although it involves several dif- ferent MutS and MutL homologs: MutSα (MSH2/MSH6) recognizes single base-base mismatches and 1–2 base insertion/deletions; MutSβ (MSH2/MSH3) recognizes insertion/ deletion mismatches containing two or more extra bases. -

WO 2016/040794 Al 17 March 2016 (17.03.2016) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2016/040794 Al 17 March 2016 (17.03.2016) P O P C T (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C12N 1/19 (2006.01) C12Q 1/02 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, C12N 15/81 (2006.01) C07K 14/47 (2006.01) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, (21) International Application Number: KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, PCT/US20 15/049674 MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, (22) International Filing Date: PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, 11 September 2015 ( 11.09.201 5) SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every (26) Publication Language: English kind of regional protection available): ARIPO (BW, GH, (30) Priority Data: GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, 62/050,045 12 September 2014 (12.09.2014) US TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, (71) Applicant: WHITEHEAD INSTITUTE FOR BIOMED¬ DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, ICAL RESEARCH [US/US]; Nine Cambridge Center, LV, MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, Cambridge, Massachusetts 02142-1479 (US). -

1 SUPPLEMENTAL DATA Figure S1. Poly I:C Induces IFN-Β Expression
SUPPLEMENTAL DATA Figure S1. Poly I:C induces IFN-β expression and signaling. Fibroblasts were incubated in media with or without Poly I:C for 24 h. RNA was isolated and processed for microarray analysis. Genes showing >2-fold up- or down-regulation compared to control fibroblasts were analyzed using Ingenuity Pathway Analysis Software (Red color, up-regulation; Green color, down-regulation). The transcripts with known gene identifiers (HUGO gene symbols) were entered into the Ingenuity Pathways Knowledge Base IPA 4.0. Each gene identifier mapped in the Ingenuity Pathways Knowledge Base was termed as a focus gene, which was overlaid into a global molecular network established from the information in the Ingenuity Pathways Knowledge Base. Each network contained a maximum of 35 focus genes. 1 Figure S2. The overlap of genes regulated by Poly I:C and by IFN. Bioinformatics analysis was conducted to generate a list of 2003 genes showing >2 fold up or down- regulation in fibroblasts treated with Poly I:C for 24 h. The overlap of this gene set with the 117 skin gene IFN Core Signature comprised of datasets of skin cells stimulated by IFN (Wong et al, 2012) was generated using Microsoft Excel. 2 Symbol Description polyIC 24h IFN 24h CXCL10 chemokine (C-X-C motif) ligand 10 129 7.14 CCL5 chemokine (C-C motif) ligand 5 118 1.12 CCL5 chemokine (C-C motif) ligand 5 115 1.01 OASL 2'-5'-oligoadenylate synthetase-like 83.3 9.52 CCL8 chemokine (C-C motif) ligand 8 78.5 3.25 IDO1 indoleamine 2,3-dioxygenase 1 76.3 3.5 IFI27 interferon, alpha-inducible -

Download (PDF)
Supplemental Table 1A Fold change Affymetrix probe Gene title Gene symbol (log2 ratio to ID control) 1424525_at gastrin releasing peptide Grp 5.80 1427747_a_at lipocalin 2 Lcn2 5.27 1449833_at small proline‐rich protein 2F Sprr2f 5.04 1450297_at interleukin 6 Il6 5.02 1450364_a_at hepatitis A virus cellular receptor 1 Havcr1 4.42 1452098_at CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) Chtf18 4.25 1419282_at chemokine (C‐C motif) ligand 12 Ccl12 4.11 serine (or cysteine) peptidase inhibitor, clade A (alpha‐1 antiproteinase, 1424758_s_at Serpina10 3.96 antitrypsin), member 10 1460227_at tissue inhibitor of metalloproteinase 1 Timp1 3.86 1419100_at serine (or cysteine) peptidase inhibitor, clade A, member 3N Serpina3n 3.59 1418626_a_at clusterin Clu 3.31 1416025_at fibrinogen, gamma polypeptide Fgg 3.25 1448756_at S100 calcium binding protein A9 (calgranulin B) S100a9 3.18 1427682_a_at early growth response 2 Egr2 3.17 1452279_at complement factor properdin Cfp 3.03 1422851_at high mobility group AT‐hook 2 Hmga2 3.01 1419579_at solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 Slc7a12 2.98 1419209_at chemokine (C‐X‐C motif) ligand 1 Cxcl1 2.92 1419764_at chitinase 3‐like 3 Chi3l3 2.91 1423954_at complement component 3 C3 2.88 1424279_at fibrinogen, alpha polypeptide Fga 2.86 1424556_at pyrroline‐5‐carboxylate reductase 1 Pycr1 2.77 1418778_at coiled‐coil domain containing 109B Ccdc109b 2.76 1422864_at runt related transcription factor 1 Runx1 2.65 1433966_x_at asparagine synthetase -

Molecular Basis for the Distinct Cellular Functions of the Lsm1-7 and Lsm2-8 Complexes
Downloaded from rnajournal.cshlp.org on October 3, 2021 - Published by Cold Spring Harbor Laboratory Press Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes Eric J. Montemayor1,2, Johanna M. Virta1, Samuel M. Hayes1, Yuichiro Nomura1, David A. Brow2, Samuel E. Butcher1 1Department of Biochemistry, University of Wisconsin-Madison, Madison, WI, USA. 2Department of Biomolecular Chemistry, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA. Correspondence should be addressed to E.J.M. ([email protected]) and S.E.B. ([email protected]). Abstract Eukaryotes possess eight highly conserved Lsm (like Sm) proteins that assemble into circular, heteroheptameric complexes, bind RNA, and direct a diverse range of biological processes. Among the many essential functions of Lsm proteins, the cytoplasmic Lsm1-7 complex initiates mRNA decay, while the nuclear Lsm2-8 complex acts as a chaperone for U6 spliceosomal RNA. It has been unclear how these complexes perform their distinct functions while differing by only one out of seven subunits. Here, we elucidate the molecular basis for Lsm-RNA recognition and present four high-resolution structures of Lsm complexes bound to RNAs. The structures of Lsm2-8 bound to RNA identify the unique 2′,3′ cyclic phosphate end of U6 as a prime determinant of specificity. In contrast, the Lsm1-7 complex strongly discriminates against cyclic phosphates and tightly binds to oligouridylate tracts with terminal purines. Lsm5 uniquely recognizes purine bases, explaining its divergent sequence relative to other Lsm subunits. Lsm1-7 loads onto RNA from the 3′ end and removal of the Lsm1 C-terminal region allows Lsm1-7 to scan along RNA, suggesting a gated mechanism for accessing internal binding sites. -

Datasheet A10199-2 Anti-LSM5 Antibody
Product datasheet Anti-LSM5 Antibody Catalog Number: A10199-2 BOSTER BIOLOGICAL TECHNOLOGY Special NO.1, International Enterprise Center, 2nd Guanshan Road, Wuhan, China Web: www.boster.com.cn Phone: +86 27 67845390 Fax: +86 27 67845390 Email: [email protected] Basic Information Product Name Anti-LSM5 Antibody Gene Name LSM5 Source Rabbit IgG Species Reactivity human, mouse Tested Application WB,IHC-P,ICC/IF,FCM,Direct ELISA Contents 500ug/ml antibody with PBS ,0.02% NaN3 , 1mg BSA and 50% glycerol. Immunogen E.coli-derived human LSM5 recombinant protein (Position: N8-V91). Purification Immunogen affinity purified. Observed MW 12KD Dilution Ratios Western blot: 1:500-2000 Immunohistochemistry in paraffin section IHC-(P): 1:50-400 Immunocytochemistry/Immunofluorescence (ICC/IF): 1:50-400 Flow cytometry (FCM): 1-3μg/1x106 cells Direct ELISA: 1:100-1000 Storage 12 months from date of receipt,-20℃ as supplied.6 months 2 to 8℃ after reconstitution. Avoid repeated freezing and thawing Background Information U6 snRNA-associated Sm-like protein LSm5 is a protein that in humans is encoded by the LSM5 gene. Sm-like proteins were identified in a variety of organisms based on sequence homology with the Sm protein family (see SNRPD2; MIM 601061). Sm-like proteins contain the Sm sequence motif, which consists of 2 regions separated by a linker of variable length that folds as a loop. The Sm-like proteins are thought to form a stable heteromer present in tri-snRNP particles, which are important for pre-mRNA splicing. Reference Anti-LSM5 Antibody被引用在0文献中。 暂无引用 FOR RESEARCH USE ONLY. NOT FOR DIAGNOSTIC AND CLINICAL USE.