Aphis Spp. 1Y.S
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Arthur Monrad Johnson Colletion of Botanical Drawings
http://oac.cdlib.org/findaid/ark:/13030/kt7489r5rb No online items Arthur Monrad Johnson colletion of botanical drawings 1914-1941 Processed by Pat L. Walter. Louise M. Darling Biomedical Library History and Special Collections Division History and Special Collections Division UCLA 12-077 Center for Health Sciences Box 951798 Los Angeles, CA 90095-1798 Phone: 310/825-6940 Fax: 310/825-0465 Email: [email protected] URL: http://www.library.ucla.edu/libraries/biomed/his/ ©2008 The Regents of the University of California. All rights reserved. Arthur Monrad Johnson colletion 48 1 of botanical drawings 1914-1941 Descriptive Summary Title: Arthur Monrad Johnson colletion of botanical drawings, Date (inclusive): 1914-1941 Collection number: 48 Creator: Johnson, Arthur Monrad 1878-1943 Extent: 3 boxes (2.5 linear feet) Repository: University of California, Los Angeles. Library. Louise M. Darling Biomedical Library History and Special Collections Division Los Angeles, California 90095-1490 Abstract: Approximately 1000 botanical drawings, most in pen and black ink on paper, of the structural parts of angiosperms and some gymnosperms, by Arthur Monrad Johnson. Many of the illustrations have been published in the author's scientific publications, such as his "Taxonomy of the Flowering Plants" and articles on the genus Saxifraga. Dr. Johnson was both a respected botanist and an accomplished artist beyond his botanical subjects. Physical location: Collection stored off-site (Southern Regional Library Facility): Advance notice required for access. Language of Material: Collection materials in English Preferred Citation [Identification of item], Arthur Monrad Johnson colletion of botanical drawings (Manuscript collection 48). Louise M. Darling Biomedical Library History and Special Collections Division, University of California, Los Angeles. -

Full of Beans: a Study on the Alignment of Two Flowering Plants Classification Systems
Full of beans: a study on the alignment of two flowering plants classification systems Yi-Yun Cheng and Bertram Ludäscher School of Information Sciences, University of Illinois at Urbana-Champaign, USA {yiyunyc2,ludaesch}@illinois.edu Abstract. Advancements in technologies such as DNA analysis have given rise to new ways in organizing organisms in biodiversity classification systems. In this paper, we examine the feasibility of aligning two classification systems for flowering plants using a logic-based, Region Connection Calculus (RCC-5) ap- proach. The older “Cronquist system” (1981) classifies plants using their mor- phological features, while the more recent Angiosperm Phylogeny Group IV (APG IV) (2016) system classifies based on many new methods including ge- nome-level analysis. In our approach, we align pairwise concepts X and Y from two taxonomies using five basic set relations: congruence (X=Y), inclusion (X>Y), inverse inclusion (X<Y), overlap (X><Y), and disjointness (X!Y). With some of the RCC-5 relationships among the Fabaceae family (beans family) and the Sapindaceae family (maple family) uncertain, we anticipate that the merging of the two classification systems will lead to numerous merged solutions, so- called possible worlds. Our research demonstrates how logic-based alignment with ambiguities can lead to multiple merged solutions, which would not have been feasible when aligning taxonomies, classifications, or other knowledge or- ganization systems (KOS) manually. We believe that this work can introduce a novel approach for aligning KOS, where merged possible worlds can serve as a minimum viable product for engaging domain experts in the loop. Keywords: taxonomy alignment, KOS alignment, interoperability 1 Introduction With the advent of large-scale technologies and datasets, it has become increasingly difficult to organize information using a stable unitary classification scheme over time. -

The Complete Chloroplast Genome Sequence of Asparagus (Asparagus Officinalis L.) and Its Phy- Logenetic Positon Within Asparagales
Central International Journal of Plant Biology & Research Bringing Excellence in Open Access Research Note *Corresponding author Wentao Sheng, Department of Biological Technology, Nanchang Normal University, Nanchang 330032, The Complete Chloroplast Jiangxi, China, Tel: 86-0791-87619332; Fax: 86-0791- 87619332; Email: Submitted: 14 September 2017 Genome Sequence of Accepted: 09 October 2017 Published: 10 October 2017 Asparagus (Asparagus ISSN: 2333-6668 Copyright © 2017 Sheng et al. officinalis L.) and its OPEN ACCESS Keywords Phylogenetic Positon within • Asparagus officinalis L • Chloroplast genome • Phylogenomic evolution Asparagales • Asparagales Wentao Sheng*, Xuewen Chai, Yousheng Rao, Xutang, Tu, and Shangguang Du Department of Biological Technology, Nanchang Normal University, China Abstract Asparagus (Asparagus officinalis L.) is a horticultural homology of medicine and food with health care. The entire chloroplast (cp) genome of asparagus was sequenced with Hiseq4000 platform. The complete cp genome maps a circular molecule of 156,699bp built with a quadripartite organization: two inverted repeats (IRs) of 26,531bp, separated by a large single copy (LSC) sequence of 84,999bp and a small single copy (SSC) sequence of 18,638bp. A total of 112 genes comprising of 78 protein-coding genes, 30 tRNAs and 4 rRNAs were successfully annotated, 17 of which included introns. The identity, number and GC content of asparagus cp genes were similar to those of other asparagus species genomes. Analysis revealed 81 simple sequence repeat (SSR) loci, most composed of A or T, contributing to a bias in base composition. A maximum likelihood phylogenomic evolution analysis showed that asparagus was closely related to Polygonatum cyrtonema that belonged to the genus Asparagales. -

Bilirubin: an Animal Pigment in the Zingiberales and Diverse Angiosperm Orders Cary L
Florida International University FIU Digital Commons FIU Electronic Theses and Dissertations University Graduate School 11-5-2010 Bilirubin: an Animal Pigment in the Zingiberales and Diverse Angiosperm Orders Cary L. Pirone Florida International University, [email protected] DOI: 10.25148/etd.FI10122201 Follow this and additional works at: https://digitalcommons.fiu.edu/etd Part of the Biochemistry Commons, and the Botany Commons Recommended Citation Pirone, Cary L., "Bilirubin: an Animal Pigment in the Zingiberales and Diverse Angiosperm Orders" (2010). FIU Electronic Theses and Dissertations. 336. https://digitalcommons.fiu.edu/etd/336 This work is brought to you for free and open access by the University Graduate School at FIU Digital Commons. It has been accepted for inclusion in FIU Electronic Theses and Dissertations by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. FLORIDA INTERNATIONAL UNIVERSITY Miami, Florida BILIRUBIN: AN ANIMAL PIGMENT IN THE ZINGIBERALES AND DIVERSE ANGIOSPERM ORDERS A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in BIOLOGY by Cary Lunsford Pirone 2010 To: Dean Kenneth G. Furton College of Arts and Sciences This dissertation, written by Cary Lunsford Pirone, and entitled Bilirubin: An Animal Pigment in the Zingiberales and Diverse Angiosperm Orders, having been approved in respect to style and intellectual content, is referred to you for judgment. We have read this dissertation and recommend that it be approved. ______________________________________ Bradley C. Bennett ______________________________________ Timothy M. Collins ______________________________________ Maureen A. Donnelly ______________________________________ John. T. Landrum ______________________________________ J. Martin Quirke ______________________________________ David W. Lee, Major Professor Date of Defense: November 5, 2010 The dissertation of Cary Lunsford Pirone is approved. -

ABSTRACTS 117 Systematics Section, BSA / ASPT / IOPB
Systematics Section, BSA / ASPT / IOPB 466 HARDY, CHRISTOPHER R.1,2*, JERROLD I DAVIS1, breeding system. This effectively reproductively isolates the species. ROBERT B. FADEN3, AND DENNIS W. STEVENSON1,2 Previous studies have provided extensive genetic, phylogenetic and 1Bailey Hortorium, Cornell University, Ithaca, NY 14853; 2New York natural selection data which allow for a rare opportunity to now Botanical Garden, Bronx, NY 10458; 3Dept. of Botany, National study and interpret ontogenetic changes as sources of evolutionary Museum of Natural History, Smithsonian Institution, Washington, novelties in floral form. Three populations of M. cardinalis and four DC 20560 populations of M. lewisii (representing both described races) were studied from initiation of floral apex to anthesis using SEM and light Phylogenetics of Cochliostema, Geogenanthus, and microscopy. Allometric analyses were conducted on data derived an undescribed genus (Commelinaceae) using from floral organs. Sympatric populations of the species from morphology and DNA sequence data from 26S, 5S- Yosemite National Park were compared. Calyces of M. lewisii initi- NTS, rbcL, and trnL-F loci ate later than those of M. cardinalis relative to the inner whorls, and sepals are taller and more acute. Relative times of initiation of phylogenetic study was conducted on a group of three small petals, sepals and pistil are similar in both species. Petal shapes dif- genera of neotropical Commelinaceae that exhibit a variety fer between species throughout development. Corolla aperture of unusual floral morphologies and habits. Morphological A shape becomes dorso-ventrally narrow during development of M. characters and DNA sequence data from plastid (rbcL, trnL-F) and lewisii, and laterally narrow in M. -

Biogeography of the Monocotyledon Astelioid Clade (Asparagales): a History of Long-Distance Dispersal and Diversification with Emerging Habitats
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2021 Biogeography of the monocotyledon astelioid clade (Asparagales): A history of long-distance dispersal and diversification with emerging habitats Birch, Joanne L ; Kocyan, Alexander Abstract: The astelioid families (Asteliaceae, Blandfordiaceae, Boryaceae, Hypoxidaceae, and Lanari- aceae) have centers of diversity in Australasia and temperate Africa, with secondary centers of diversity in Afromontane Africa, Asia, and Pacific Islands. The global distribution of these families makes this an excellent lineage to test if current distribution patterns are the result of vicariance or long-distance dispersal and to evaluate the roles of tertiary climatic and geological drivers in lineage diversification. Sequence data were generated from five chloroplast regions (petL-psbE, rbcL, rps16-trnK, trnL-trnLF, trnS-trnSG) for 104 ingroup species sampled across global diversity. The astelioid phylogeny was inferred using maximum parsimony, maximum likelihood, and Bayesian inference methods. Divergence dates were estimated with a relaxed clock applied in BEAST. Ancestral ranges were reconstructed in ’BioGeoBEARS’ applying the corrected Akaike information criterion to test for the best-fit biogeographic model. Diver- sification rates were estimated in Bayesian Analysis of Macroevolutionary Mixtures [BAMM]. Astelioid relationships were inferred as Boryaceae(Blandfordiaceae(Asteliaceae(Hypoxidaceae plus Lanariaceae))). The crown astelioid node was dated to the Late Cretaceous (75.2 million years; 95% highest posterior densities interval 61.0-90.0 million years) with an inferred Eastern Gondwanan origin. However, aste- lioid speciation events have not been shaped by Gondwanan vicariance. Rather long-distance dispersal since the Eocene is inferred to account for current distributions. -

BOT 5725-Taxonomy of Vascular Plants
BOT 5725 Taxonomy of Vascular Plants Spring 2018 Instructors: Doug Soltis (301 Dickinson; phone: 273-1963; e-mail: [email protected]) Pam Soltis (301 Dickinson; phone: 273-1964; e-mail: [email protected]) Nico Cellinese (379 Dickinson; phone: 273-1979; e-mail: [email protected]) Emily Sessa (521 Bartram; phone: 392-1098; e-mail: [email protected]) Ryan Folk (359 Dickinson; phone: 330-801-3078; email: [email protected]) Office Hours: By appointment Credits: 4 Herbarium: 379 Dickinson Hall: phone: 273-1990. Library is open between 9:00 am and 5:00 pm, but plant collection usage restricted to faculty (or graduate students with approved systematic research projects). Lecture: Tuesday & Thursday, periods 6-9 (12:50-4:55pm); Rolfs Hall room 105. Textbook (required): Christenhusz et al. (2017) Plants of the World. Plus additional readings to be provided as PDFs on Canvas. Laboratory tools: 2 dissecting needles, package of razor blades (or scalpel), tweezers, 10X hand-lens. Grading: One mid-term exam (45% of final grade), one final exam (45% of final exam), class participation (10% of final grade). Grade based on total number of points, with 90% or above an “A”, 89-80% “B”, 79-70% “C”, 69-60% “D”, and below failing; plus and minus grades will be used. Exams will be based on lecture and laboratory material. Outline of lectures and labs Spring 2017 Week 1: 9 Jan PS Introduction: What is taxonomy? Basic principles, digital resources 11 Jan NC 12:50-1:40pm (Rolfs): Field and herbarium methods; identification and introduction to nomenclature 2pm (Dickinson): Herbarium Tour Week 2: 16 Jan NC Species concepts, speciation 18 Jan PS Phylogenetics Week 3: 23 Jan PS Phylogenetics, cont. -

Elatinaceae Are Sister to Malpighiaceae; Peridiscaceae Belong to Saxifragales
Elatinaceae are Sister to Malpighiaceae; Peridiscaceae Belong to Saxifragales The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Davis, Charles C., and Mark W. Chase. 2004. Elatinaceae are sister to Malpighiaceae; Peridiscaceae belong to Saxifragales. American Journal of Botany 91(2): 262-273. Published Version http://dx.doi.org/10.3732/ajb.91.2.262 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:2666728 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA American Journal of Botany 91(2): 262±273. 2004. ELATINACEAE ARE SISTER TO MALPIGHIACEAE; PERIDISCACEAE BELONG TO SAXIFRAGALES1 CHARLES C. DAVIS2,4 AND MARK W. C HASE3 2Department of Ecology and Evolutionary Biology, University of Michigan Herbarium, 3600 Varsity Drive, Ann Arbor, Michigan 48108-2287 USA; and 3Molecular Systematics Section, Jodrell Laboratory, Royal Botanic Gardens, Kew, Richmond, Surrey TW9 3DS UK Phylogenetic data from plastid (ndhF and rbcL) and nuclear (PHYC) genes indicate that, within the order Malpighiales, Elatinaceae are strongly supported as sister to Malpighiaceae. There are several putative morphological synapomorphies for this clade; most notably, they both have a base chromosome number of X 5 6 (or some multiple of three or six), opposite or whorled leaves with stipules, unicellular hairs (also uniseriate in some Elatinaceae), multicellular glands on the leaves, and resin (Elatinacae) or latex (Malpighiaceae). -

Lesson 3 RECOGNISING PLANT FAMILIES and IDENTIFYING PLANTS Aim Distinguish Between Different Plant Families and to Become Profi
Lesson 3 RECOGNISING PLANT FAMILIES AND IDENTIFYING PLANTS Aim Distinguish between different plant families and to become proficient at identifying plants. The best way to build your ability to identify plants is by working with or handling a variety of different plants on a daily basis. The first ten plant names you learn are always much more difficult than the next ninety. Similarly, the first 100 names are always much more difficult than the next 900. If you plan to be a skilled gardener, landscape designer, or horticulturist: you need to learn to identify hundreds of different plants. As you have seen earlier; there is a system in identifying plant names. BECOME FAMILIAR WITH PLANT FAMILIES If you can get to know the way the system works, and the broad categories, the whole thing starts to make a great deal more sense. Each new name you confront is able to be associated with things and remembered more easily. FOR EXAMPLE: “When I see a plant with a daisy flower, I immediately know that it is in the Asteraceae family. Even if the genus is new to me, I will be more likely to remember it because I’m not only thinking: This is the genus of this new plant , but I am also thinking: This new genus is in the Asteraceae family. In essence, my brain is registering two pieces of information instead of one and that doubles the likelihood of me remembering the plant. BECOME FAMILIAR WITH LATIN Plant naming is based on the ancient Roman language of Latin. -
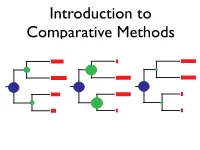
BM CC EB What Can We Learn from a Tree?
Introduction to Comparative Methods BM CC EB What can we learn from a tree? Net diversification (r) Relative extinction (ε) Peridiscaceae Peridiscaceae yllaceae yllaceae h h atop atop Proteaceae Proteaceae r r Ce Ce Tr oc T ho r M M o de c y y H H C C h r r e e a D D a o o nd o e e G G a a m m t t a a d r P r P h h e e u u c c p A p A r e a a e e a a a c a c n n a i B i B h h n d m d l m a l a m m e a e a e e t t n n c u u n n i i d i i e e e e o n o n p n p n a e a S e e S e e n n x x i i r c c a a n o n o p p h g e h g ae e l r a l r a a a a a a i i a a a e a e i b i b h y d c h d c i y i c a a c x c x c c G I a G I a n c n c c c y l y l t a a t a a e e e e e i l c i l c m l m l e c e c f a e a a f a e a a l r r l c c a i i r l e t e t a a r l a a e e u u u u o a o a a a c a c a a l a l e e e b b a a a a e e c e e c a a s c s c c e l c e l e e g e g e a a a a e e n n s e e s e e e e a a a P a P e e N N u u u S u S a e a e a a e e c c l n a l n e e a e e a e a e a e a e r a r a c c C i C i R R a e a e a e a e r c r c A A a d a a d a e i e i phanopetalaceae s r e ph s r e a a s e c s e c e e u u b a a b a e e P P r r l l e n e a a a a m m entho e e e Ha a H o a c r e c r e nt B B e p e e e c e e c a c c h e a p a a p a lo lo l l a a e s o t e s i a r a i a r r r r r a n e a n e a b a l b t a t gaceae e g e ceae a c a s c s a z e z M i a e M i a c a d e a d e ae e ae r e r a e a e a a a c c ce r e r L L i i ac a Vitaceae Vi r r C C e e ta v e v e a a c a a e ea p e ap c a c a e a P P e e l l e Ge G e e ae a t t e e p p r r ce c an u an -

Supplementary Information
Supporting Information for Cornwell et al. 2014. Functional distinctiveness of major 1 plant lineages. doi: 10.1111/1365-2745.12208. SUPPORTING INFORMATION Summary In this Supporting Information, we report additional methods details with respect to the trait data, the climate data, and the analysis. In addition, to more fully explain the method, we include several additional figures. We show the top-five lineages, including the population of lineages from which they were selected, for five important traits (Figure S1 on five separate pages); the bivariate distribution of three selected clades with respect to SLA and leaf N, components of the leaf economic spectrum (Figure S2), the geographic distribution of the clades where this proved useful for interpretation (Figure S3); the procedure for selecting the top six nodes in the leaf N trait, to illustrate the internal behavior of our new comparative method, especially the relative contribution of components of extremeness and the sample size weighting to the results (Figure S4). Finally, we provide references for data used in analyses. SUPPORTING METHODS TRAITS DATABASE We compiled a database for five plant functional traits. Each of these traits is the result of a separate research initiative in which data were gathered directly from researchers leading those efforts and/or the literature; in most, but not all cases, these data have been published elsewhere. Detailed methods for data collection and assembly for each trait are available in the original publications; further data were added for some traits from the primary literature (for references see description of individual traits and Supporting References below). For our compilation, all data were brought to common units for a given trait and thoroughly error checked. -

Saxifragaceae Sensu Lato (DNA Sequencing/Evolution/Systematics) DOUGLAS E
Proc. Nati. Acad. Sci. USA Vol. 87, pp. 4640-4644, June 1990 Evolution rbcL sequence divergence and phylogenetic relationships in Saxifragaceae sensu lato (DNA sequencing/evolution/systematics) DOUGLAS E. SOLTISt, PAMELA S. SOLTISt, MICHAEL T. CLEGGt, AND MARY DURBINt tDepartment of Botany, Washington State University, Pullman, WA 99164; and tDepartment of Botany and Plant Sciences, University of California, Riverside, CA 92521 Communicated by R. W. Allard, March 19, 1990 (received for review January 29, 1990) ABSTRACT Phylogenetic relationships are often poorly quenced and analyses to date indicate that it is reliable for understood at higher taxonomic levels (family and above) phylogenetic analysis at higher taxonomic levels, (ii) rbcL is despite intensive morphological analysis. An excellent example a large gene [>1400 base pairs (bp)] that provides numerous is Saxifragaceae sensu lato, which represents one of the major characters (bp) for phylogenetic studies, and (iii) the rate of phylogenetic problems in angiosperms at higher taxonomic evolution of rbcL is appropriate for addressing questions of levels. As originally defined, the family is a heterogeneous angiosperm phylogeny at the familial level or higher. assemblage of herbaceous and woody taxa comprising 15 We used rbcL sequence data to analyze phylogenetic subfamilies. Although more recent classifications fundamen- relationships in a particularly problematic group-Engler's tally modified this scheme, little agreement exists regarding the (8) broadly defined family Saxifragaceae (Saxifragaceae circumscription, taxonomic rank, or relationships of these sensu lato). Based on morphological analyses, the group is subfamilies. The recurrent discrepancies in taxonomic treat- almost impossible to distinguish or characterize clearly and ments of the Saxifragaceae prompted an investigation of the taxonomic problems at higher power of chloroplast gene sequences to resolve phylogenetic represents one of the greatest relationships within this family and between the Saxifragaceae levels in the angiosperms (9, 10).