Eph Receptor B1 (EPHB1) (NM 004441) Human Tagged ORF Clone Product Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pancancer Progression Human Vjune2017
Gene Symbol Accession Alias/Prev Symbol Official Full Name AAMP NM_001087.3 - angio-associated, migratory cell protein ABI3BP NM_015429.3 NESHBP|TARSH ABI family, member 3 (NESH) binding protein ACHE NM_000665.3 ACEE|ARACHE|N-ACHE|YT acetylcholinesterase ACTG2 NM_001615.3 ACT|ACTA3|ACTE|ACTL3|ACTSG actin, gamma 2, smooth muscle, enteric ACVR1 NM_001105.2 ACTRI|ACVR1A|ACVRLK2|ALK2|FOP|SKR1|TSRI activin A receptor, type I ACVR1C NM_145259.2 ACVRLK7|ALK7 activin A receptor, type IC ACVRL1 NM_000020.1 ACVRLK1|ALK-1|ALK1|HHT|HHT2|ORW2|SKR3|TSR-I activin A receptor type II-like 1 ADAM15 NM_207195.1 MDC15 ADAM metallopeptidase domain 15 ADAM17 NM_003183.4 ADAM18|CD156B|CSVP|NISBD|TACE ADAM metallopeptidase domain 17 ADAM28 NM_014265.4 ADAM 28|ADAM23|MDC-L|MDC-Lm|MDC-Ls|MDCL|eMDC II|eMDCII ADAM metallopeptidase domain 28 ADAM8 NM_001109.4 CD156|MS2 ADAM metallopeptidase domain 8 ADAM9 NM_001005845.1 CORD9|MCMP|MDC9|Mltng ADAM metallopeptidase domain 9 ADAMTS1 NM_006988.3 C3-C5|METH1 ADAM metallopeptidase with thrombospondin type 1 motif, 1 ADAMTS12 NM_030955.2 PRO4389 ADAM metallopeptidase with thrombospondin type 1 motif, 12 ADAMTS8 NM_007037.4 ADAM-TS8|METH2 ADAM metallopeptidase with thrombospondin type 1 motif, 8 ADAP1 NM_006869.2 CENTA1|GCS1L|p42IP4 ArfGAP with dual PH domains 1 ADD1 NM_001119.4 ADDA adducin 1 (alpha) ADM2 NM_001253845.1 AM2|dJ579N16.4 adrenomedullin 2 ADRA2B NM_000682.4 ADRA2L1|ADRA2RL1|ADRARL1|ALPHA2BAR|alpha-2BAR adrenoceptor alpha 2B AEBP1 NM_001129.3 ACLP AE binding protein 1 AGGF1 NM_018046.3 GPATC7|GPATCH7|HSU84971|HUS84971|VG5Q -

Coexistence of Eph Receptor B1 and Ephrin B2 in Port-Wine Stain Endothelial Progenitor Cells Contributes to Clinicopathological Vasculature Dilatation
Coexistence of Eph receptor B1 and ephrin B2 in port-wine stain endothelial progenitor cells contributes to clinicopathological vasculature dilatation W. Tan iD ,1 J. Wang,1,2 F. Zhou,1,2 L. Gao,1,3 R. Yin iD ,1,4 H. Liu,5 A. Sukanthanag,1 G. Wang,3 M.C. Mihm Jr.,6 D.-B. Chen7 and J.S. Nelson1,8 1Department of Surgery, Beckman Laser Institute and Medical Clinic; 7Department of Obstetrics and Gynecology; and 8Department of Biomedical Engineering, University of California, Irvine, Irvine, CA, U.S.A. 2The Third Xiangya Hospital, Xiangya School of Medicine, Central South University, Changsha, Hunan 412000, China 3Department of Dermatology, Xijing Hospital, Fourth Military Medical University, Xi’an, 710032, China 4Department of Dermatology, The Second Hospital of Shanxi Medical University, Taiyuan 030001, China 5Shandong Provincial Institute of Dermatology and Venereology, Jinan, Shandong 250022, China 6Department of Dermatology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, U.S.A. Summary Background Port-wine stain (PWS) is a vascular malformation characterized by progressive dilatation of postcapillary venules, but the molecular pathogenesis remains obscure. Objectives To illustrate that PWS endothelial cells (ECs) present a unique molecular phenotype that leads to pathoanatomical PWS vasculatures. Methods Immunohistochemistry and transmission electron microscopy were used to characterize the ultrastructure and molecular phenotypes of PWS blood vessels. Primary culture of human dermal microvascular endothelial cells and in vitro tube formation assay were used for confirmative functional studies. Results Multiple clinicopathological features of PWS blood vessels during the development and progression of the disease were shown. There were no normal arterioles and venules observed phenotypically and morphologically in PWS skin; arterioles and venules both showed differentiation impairments, resulting in a reduction of arteriole- like vasculatures and defects in capillary loop formation in PWS lesions. -

Supplementary Table 1. in Vitro Side Effect Profiling Study for LDN/OSU-0212320. Neurotransmitter Related Steroids
Supplementary Table 1. In vitro side effect profiling study for LDN/OSU-0212320. Percent Inhibition Receptor 10 µM Neurotransmitter Related Adenosine, Non-selective 7.29% Adrenergic, Alpha 1, Non-selective 24.98% Adrenergic, Alpha 2, Non-selective 27.18% Adrenergic, Beta, Non-selective -20.94% Dopamine Transporter 8.69% Dopamine, D1 (h) 8.48% Dopamine, D2s (h) 4.06% GABA A, Agonist Site -16.15% GABA A, BDZ, alpha 1 site 12.73% GABA-B 13.60% Glutamate, AMPA Site (Ionotropic) 12.06% Glutamate, Kainate Site (Ionotropic) -1.03% Glutamate, NMDA Agonist Site (Ionotropic) 0.12% Glutamate, NMDA, Glycine (Stry-insens Site) 9.84% (Ionotropic) Glycine, Strychnine-sensitive 0.99% Histamine, H1 -5.54% Histamine, H2 16.54% Histamine, H3 4.80% Melatonin, Non-selective -5.54% Muscarinic, M1 (hr) -1.88% Muscarinic, M2 (h) 0.82% Muscarinic, Non-selective, Central 29.04% Muscarinic, Non-selective, Peripheral 0.29% Nicotinic, Neuronal (-BnTx insensitive) 7.85% Norepinephrine Transporter 2.87% Opioid, Non-selective -0.09% Opioid, Orphanin, ORL1 (h) 11.55% Serotonin Transporter -3.02% Serotonin, Non-selective 26.33% Sigma, Non-Selective 10.19% Steroids Estrogen 11.16% 1 Percent Inhibition Receptor 10 µM Testosterone (cytosolic) (h) 12.50% Ion Channels Calcium Channel, Type L (Dihydropyridine Site) 43.18% Calcium Channel, Type N 4.15% Potassium Channel, ATP-Sensitive -4.05% Potassium Channel, Ca2+ Act., VI 17.80% Potassium Channel, I(Kr) (hERG) (h) -6.44% Sodium, Site 2 -0.39% Second Messengers Nitric Oxide, NOS (Neuronal-Binding) -17.09% Prostaglandins Leukotriene, -

Beyond Traditional Morphological Characterization of Lung
Cancers 2020 S1 of S15 Beyond Traditional Morphological Characterization of Lung Neuroendocrine Neoplasms: In Silico Study of Next-Generation Sequencing Mutations Analysis across the Four World Health Organization Defined Groups Giovanni Centonze, Davide Biganzoli, Natalie Prinzi, Sara Pusceddu, Alessandro Mangogna, Elena Tamborini, Federica Perrone, Adele Busico, Vincenzo Lagano, Laura Cattaneo, Gabriella Sozzi, Luca Roz, Elia Biganzoli and Massimo Milione Table S1. Genes Frequently mutated in Typical Carcinoids (TCs). Mutation Original Entrez Gene Gene Rate % eukaryotic translation initiation factor 1A X-linked [Source: HGNC 4.84 EIF1AX 1964 EIF1AX Symbol; Acc: HGNC: 3250] AT-rich interaction domain 1A [Source: HGNC Symbol;Acc: HGNC: 4.71 ARID1A 8289 ARID1A 11110] LDL receptor related protein 1B [Source: HGNC Symbol; Acc: 4.35 LRP1B 53353 LRP1B HGNC: 6693] 3.53 NF1 4763 NF1 neurofibromin 1 [Source: HGNC Symbol;Acc: HGNC: 7765] DS cell adhesion molecule like 1 [Source: HGNC Symbol; Acc: 2.90 DSCAML1 57453 DSCAML1 HGNC: 14656] 2.90 DST 667 DST dystonin [Source: HGNC Symbol;Acc: HGNC: 1090] FA complementation group D2 [Source: HGNC Symbol; Acc: 2.90 FANCD2 2177 FANCD2 HGNC: 3585] piccolo presynaptic cytomatrix protein [Source: HGNC Symbol; Acc: 2.90 PCLO 27445 PCLO HGNC: 13406] erb-b2 receptor tyrosine kinase 2 [Source: HGNC Symbol; Acc: 2.44 ERBB2 2064 ERBB2 HGNC: 3430] BRCA1 associated protein 1 [Source: HGNC Symbol; Acc: HGNC: 2.35 BAP1 8314 BAP1 950] capicua transcriptional repressor [Source: HGNC Symbol; Acc: 2.35 CIC 23152 CIC HGNC: -

Description: Uniprot:P54762 Alternative Names: Specificity
TA8145 Phospho-EPHB1/2/3/4 (Tyr600/Tyr602/Tyr614/Tyr596) Antibody Order 021-34695924 [email protected] Support 400-6123-828 50ul [email protected] 100 uL √ √ Web www.ab-mart.com.cn Description: Receptor tyrosine kinase which binds promiscuously transmembrane ephrin-B family ligands residing on adjacent cells, leading to contact-dependent bidirectional signaling into neighboring cells. The signaling pathway downstream of the receptor is referred to as forward signaling while the signaling pathway downstream of the ephrin ligand is referred to as reverse signaling. Cognate/functional ephrin ligands for this receptor include EFNB1, EFNB2 and EFNB3. During nervous system development, regulates retinal axon guidance redirecting ipsilaterally ventrotemporal retinal ganglion cells axons at the optic chiasm midline. This probably requires repulsive interaction with EFNB2. In the adult nervous system together with EFNB3, regulates chemotaxis, proliferation and polarity of the hippocampus neural progenitors. In addition to its role in axon guidance plays also an important redundant role with other ephrin-B receptors in development and maturation of dendritic spines and synapse formation. May also regulate angiogenesis. More generally, may play a role in targeted cell migration and adhesion. Upon activation by EFNB1 and probably other ephrin-B ligands activates the MAPK/ERK and the JNK signaling cascades to regulate cell migration and adhesion respectively. Involved in the maintenance of the pool of satellite cells (muscle stem cells) by promoting -

Gene Symbol Accession Alias/Prev Symbol Official Full Name AAK1 NM 014911.2 KIAA1048, Dkfzp686k16132 AP2 Associated Kinase 1
Gene Symbol Accession Alias/Prev Symbol Official Full Name AAK1 NM_014911.2 KIAA1048, DKFZp686K16132 AP2 associated kinase 1 (AAK1) AATK NM_001080395.2 AATYK, AATYK1, KIAA0641, LMR1, LMTK1, p35BP apoptosis-associated tyrosine kinase (AATK) ABL1 NM_007313.2 ABL, JTK7, c-ABL, p150 v-abl Abelson murine leukemia viral oncogene homolog 1 (ABL1) ABL2 NM_007314.3 ABLL, ARG v-abl Abelson murine leukemia viral oncogene homolog 2 (arg, Abelson-related gene) (ABL2) ACVR1 NM_001105.2 ACVRLK2, SKR1, ALK2, ACVR1A activin A receptor ACVR1B NM_004302.3 ACVRLK4, ALK4, SKR2, ActRIB activin A receptor, type IB (ACVR1B) ACVR1C NM_145259.2 ACVRLK7, ALK7 activin A receptor, type IC (ACVR1C) ACVR2A NM_001616.3 ACVR2, ACTRII activin A receptor ACVR2B NM_001106.2 ActR-IIB activin A receptor ACVRL1 NM_000020.1 ACVRLK1, ORW2, HHT2, ALK1, HHT activin A receptor type II-like 1 (ACVRL1) ADCK1 NM_020421.2 FLJ39600 aarF domain containing kinase 1 (ADCK1) ADCK2 NM_052853.3 MGC20727 aarF domain containing kinase 2 (ADCK2) ADCK3 NM_020247.3 CABC1, COQ8, SCAR9 chaperone, ABC1 activity of bc1 complex like (S. pombe) (CABC1) ADCK4 NM_024876.3 aarF domain containing kinase 4 (ADCK4) ADCK5 NM_174922.3 FLJ35454 aarF domain containing kinase 5 (ADCK5) ADRBK1 NM_001619.2 GRK2, BARK1 adrenergic, beta, receptor kinase 1 (ADRBK1) ADRBK2 NM_005160.2 GRK3, BARK2 adrenergic, beta, receptor kinase 2 (ADRBK2) AKT1 NM_001014431.1 RAC, PKB, PRKBA, AKT v-akt murine thymoma viral oncogene homolog 1 (AKT1) AKT2 NM_001626.2 v-akt murine thymoma viral oncogene homolog 2 (AKT2) AKT3 NM_181690.1 -

Novel Tumor-Specific Mutations in Receptor Tyrosine Kinase Subdomain IX Significantly Reduce Extracellular Signal-Regulated Kinase Activity
ANTICANCER RESEARCH 36: 2733-2744 (2016) Novel Tumor-specific Mutations in Receptor Tyrosine Kinase Subdomain IX Significantly Reduce Extracellular Signal-regulated Kinase Activity MASAKUNI SERIZAWA1, MASATOSHI KUSUHARA1,2, SUMIKO OHNAMI3, TAKESHI NAGASHIMA3,4, YUJI SHIMODA3,4, KEIICHI OHSHIMA5, TOHRU MOCHIZUKI5, KENICHI URAKAMI3 and KEN YAMAGUCHI6 1Drug Discovery and Development Division, 2Region Resources Division, 3Cancer Diagnostics Division, and 5Medical Genetics Division, Shizuoka Cancer Center Research Institute, Shizuoka, Japan; 4SRL, Inc., Tokyo, Japan; 6Shizuoka Cancer Center Hospital and Research Institute, Shizuoka, Japan Abstract. Background/Aim: The identification of additional targeted cancer therapeutics against tumors with oncogenic therapeutic targets by clinical molecular profiling is necessary genetic alterations (4, 5). However, only a handful of patients to expand the range of molecular-targeted cancer with cancer actually benefit from effective molecular- therapeutics. This study aimed to identify novel functional targeted cancer therapeutics. In order to expand the range of tumor-specific single nucleotide variants (SNVs) in the kinase molecular-targeted cancer therapeutics, the identification of domain of receptor tyrosine kinases (RTKs), from whole-exome additional therapeutic targets through molecular profiling of sequencing (WES) data. Materials and Methods: SNVs were each patient with cancer is urgently needed (6). Therefore, selected from WES data of multiple cancer types using both the Shizuoka Cancer Center -

Kinome Expression Profiling to Target New Therapeutic Avenues in Multiple Myeloma
Plasma Cell DIsorders SUPPLEMENTARY APPENDIX Kinome expression profiling to target new therapeutic avenues in multiple myeloma Hugues de Boussac, 1 Angélique Bruyer, 1 Michel Jourdan, 1 Anke Maes, 2 Nicolas Robert, 3 Claire Gourzones, 1 Laure Vincent, 4 Anja Seckinger, 5,6 Guillaume Cartron, 4,7,8 Dirk Hose, 5,6 Elke De Bruyne, 2 Alboukadel Kassambara, 1 Philippe Pasero 1 and Jérôme Moreaux 1,3,8 1IGH, CNRS, Université de Montpellier, Montpellier, France; 2Department of Hematology and Immunology, Myeloma Center Brussels, Vrije Universiteit Brussel, Brussels, Belgium; 3CHU Montpellier, Laboratory for Monitoring Innovative Therapies, Department of Biologi - cal Hematology, Montpellier, France; 4CHU Montpellier, Department of Clinical Hematology, Montpellier, France; 5Medizinische Klinik und Poliklinik V, Universitätsklinikum Heidelberg, Heidelberg, Germany; 6Nationales Centrum für Tumorerkrankungen, Heidelberg , Ger - many; 7Université de Montpellier, UMR CNRS 5235, Montpellier, France and 8 Université de Montpellier, UFR de Médecine, Montpel - lier, France ©2020 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2018.208306 Received: October 5, 2018. Accepted: July 5, 2019. Pre-published: July 9, 2019. Correspondence: JEROME MOREAUX - [email protected] Supplementary experiment procedures Kinome Index A list of 661 genes of kinases or kinases related have been extracted from literature9, and challenged in the HM cohort for OS prognostic values The prognostic value of each of the genes was computed using maximally selected rank test from R package MaxStat. After Benjamini Hochberg multiple testing correction a list of 104 significant prognostic genes has been extracted. This second list has then been challenged for similar prognosis value in the UAMS-TT2 validation cohort. -

Blood Vessels Guide Schwann Cell Migration in the Adult Demyelinated
bioRxiv preprint doi: https://doi.org/10.1101/498261; this version posted December 16, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Title: Blood vessels guide Schwann cell migration in the adult demyelinated CNS through Eph/ephrin signaling Authors and affiliations: Beatriz Garcia-Diaz*1,2, Corinne Bachelin2, Fanny Coulpier3, Gaspard Gerschenfeld3, Cyrille Deboux2, Violetta Zujovic2, Patrick Charnay3, Piotr Topilko3, Anne Baron-Van Evercooren*2. 1Unidad de Gestión Clínica de Neurociencias, IBIMA, Hospital Regional Universitario de Málaga, Universidad de Málaga, Spain. 2Institut du Cerveau et de la Moelle Epinière-Groupe Hospitalier Pitié-Salpêtrière, INSERM, U1127, CNRS, UMR 7225, F-75013 Paris; Sorbonne Universités, Université Pierre et Marie Curie Paris 06, UM-75, F-75005 Paris, France. 3Ecole normale supérieure, PSL Research University, CNRS, Inserm, Institut de Biologie de l’Ecole normale supérieure (IBENS), F-75005 Paris, France Conflict of interest : The authors have declared that no conflict of interest exists. *Corresponding authors Beatriz Garcia Diaz, PhD Anne Baron Van-Evercooren, PhD Brain and Spine Institute (ICM) Brain and Spine Institute (ICM) ICM Building, 5th floor, 5.031 ICM Building, 5th floor, 5.031 47 bd de l'Hôpital 47 bd de l'Hôpital Paris 75013 Paris 75013 France France Tel : +33 (0)1 57 27 4170 Tel : +33 (0) 01 57 27 41 23 e-mail: [email protected] e-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/498261; this version posted December 16, 2018. -
List of Actionable Genes and Potential Matched Therapy
Supplementary Table S1: List of actionable genes and potential matched therapy Gene Level Potential Matched Therapy AKT1 3 AKT inhibitor ALK fusion 1 ALK inhibitor ARAF 3 Sorafenib BRAF V600E 2A BRAF inhibitors BRCA1/2 loss 2B PARP Inhibitor therapy CD74-NRG 4 Anti-HER3 Monoclonal Antibody CDK4 AMP 2B CDK 4/6 inhibitors CDKN2A loss 4 CDK4/6 inhibitors c-KIT 2B Imatinib EGFR 1 EGFR Tyrosine kinase inhibitors ERBB2 mutation 3 HER2 Tyrosine kinase inhibitors ERBB2 Amplification 2B Pan-HER TKI e.g. Dacomitinib, anti-HER2 directed therapy e.g. Trastuzumab ERBB3 4 ERBB3 kinase inhibitor, anti-HER3 monoclonal antibody FGFR1 3 FGFR1 inhibitors FGFR2 3 FGFR2 Inhibitors FGFR3 3 FGFR3 inhibitors FGFR3-TACC3 fusion 3 FGFR3 inhibitors GNAS 4 MEK Inhibitors HRAS 4 MEK Inhibitors KRAS 4 MEK inhibitors MAP2K1 3 MEK Inhibitors MDM2 Amplification 4 MDM2 inhibitors MET exon 14 alteration 2B Cabozantinib, Crizotinib MET Amplification 2B Cabozantinib, Crizotinib NF1 loss 4 MEK Inhibitors NRAS 3 MEK Inhibitors PIK3CA 3 PI3K, Akt , mTOR inhibitors PTEN loss 4 PI3K, Akt , mTOR inhibitors RAF1 4 Sorafenib RET fusion 2A RET inhibitors ROS1 fusion 1 ROS1 inhibitors TSC1/TSC2 loss 2B mTOR inhibitors Supplementary Table S2: List of cooccurring Level 1-4 mutations Cooccurring Level 1 Level 2A Level 2B Level 3 Level 4 N= Level 1+2A EGFR L747_P753delinsS MET AMP CDK4 AMP CDKN2A loss 1 EGFR E746_A750del MET AMP PIK3CA E545K 1 EGFR L858R MET AMP CDKN2A loss 1 MDM2 AMP EGFR L858R MET AMP CDKN2A loss 1 EGFR L858R MET AMP BRAF D594V 1 EGFR L858R MET AMP CDKN2A loss 1 EGFR -
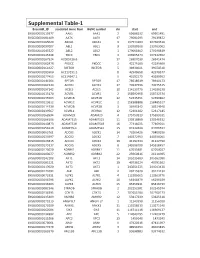
Supplemental Table-1
Supplemental Table-1 Ensembl_ID Associated Gene Nam HGNC symbol chr start end ENSG00000115977 AAK1 AAK1 2 69688532 69901481 ENSG00000181409 AATK AATK 17 79091095 79139872 ENSG00000165029 ABCA1 ABCA1 9 107543283 107690518 ENSG00000097007 ABL1 ABL1 9 133589268 133763062 ENSG00000143322 ABL2 ABL2 1 179068462 179198819 ENSG00000145348 TBCK TBCK 4 106965474 107242652 ENSG00000167524 AC005726.6 17 26907030 26941474 ENSG00000162878 PKDCC PKDCC 2 42275160 42285668 ENSG00000164327 RICTOR RICTOR 5 38938021 39074510 ENSG00000185900 AC113191.1 8 42948658 42978577 ENSG00000177453 AC114947.1 5 43192173 43280952 ENSG00000141564 RPTOR RPTOR 17 78518619 78940173 ENSG00000161533 ACOX1 ACOX1 17 73937596 73975515 ENSG00000197142 ACSL5 ACSL5 10 114133776 114188138 ENSG00000115170 ACVR1 ACVR1 2 158592958 158732374 ENSG00000135503 ACVR1B ACVR1B 12 52345451 52390862 ENSG00000123612 ACVR1C ACVR1C 2 158388888 158485517 ENSG00000114739 ACVR2B ACVR2B 3 38495342 38524948 ENSG00000139567 ACVRL1 ACVRL1 12 52301202 52317145 ENSG00000168594 ADAM29 ADAM29 4 175750819 175899331 ENSG00000166106 ADAMTS15 ADAMTS15 11 130318869 130346532 ENSG00000140873 ADAMTS18 ADAMTS18 16 77316025 77469011 ENSG00000156218 ADAMTSL3 ADAMTSL3 15 84322838 84708594 ENSG00000063761 ADCK1 ADCK1 14 78266426 78400296 ENSG00000133597 ADCK2 ADCK2 7 140372953 140396061 ENSG00000123815 ADCK4 ADCK4 19 41197434 41222790 ENSG00000173137 ADCK5 ADCK5 8 145596790 145618457 ENSG00000173020 ADRBK1 ADRBK1 11 67033881 67054027 ENSG00000100077 ADRBK2 ADRBK2 22 25960816 26118985 ENSG00000142208 AKT1 AKT1 14 105235689 -

Eph-Ephrin Signaling Modulated by Polymerization and Condensation of Receptors
Eph-ephrin signaling modulated by polymerization and condensation of receptors Samuel Ojosnegrosa,b,c,d,1,3, Francesco Cutralea,c,d,1, Daniel Rodrígueza,e, Jason J. Otterstromf, Chi Li Chiug, Verónica Hortigüelah,i, Carolina Tarantinob, Anna Seriolab, Stephen Mieruszynskij, Elena Martínezh,i,k, Melike Lakadamyalif, Angel Rayab,i,l,2, and Scott E. Fraserb,2,3 aBiology Division, California Institute of Technology, Pasadena, CA 91125; bCenter of Regenerative Medicine in Barcelona, Hospital Duran i Reynals, Hospitalet de Llobregat, 08908 Barcelona, Spain; cTranslational Imaging Center, University of Southern California, Los Angeles, CA 90089; dDepartment of Biological Sciences, Molecular and Computational Biology Section, University of Southern California, Los Angeles, CA 90089; eLaboratory of Theoretical & Applied Mechanics, Department of Mechanical Engineering, Universidade Federal Fluminense, Niterói, RJ 24210-240, Brazil; fICFO-The Institute of Photonic Sciences, The Barcelona Institute of Science and Technology, 08860 Castelldefels (Barcelona), Spain; gCenter for Applied Molecular Medicine, University of Southern California, Los Angeles, CA 90033; hBiomimetic Systems for Cell Engineering Group, Institute for Bioengineering of Catalonia, 08028 Barcelona, Spain; iBiomedical Research Networking Center in Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN), 28029 Madrid, Spain; jEuropean Molecular Biology Laboratory Australia, Australian Regenerative Medicine Institute, Monash University, Clayton, VIC 3800, Australia; kElectronics Department, University of Barcelona, 08028 Barcelona, Spain; and lInstitució Catalana de Recerca i Estudis Avançats, 08010 Barcelona, Spain Edited by Harry B. Gray, California Institute of Technology, Pasadena, CA, and approved October 31, 2017 (received for review August 1, 2017) Eph receptor signaling plays key roles in vertebrate tissue boundary interpreted by the Eph receptor into proportional responses is largely formation, axonal pathfinding, and stem cell regeneration by steering unknown.