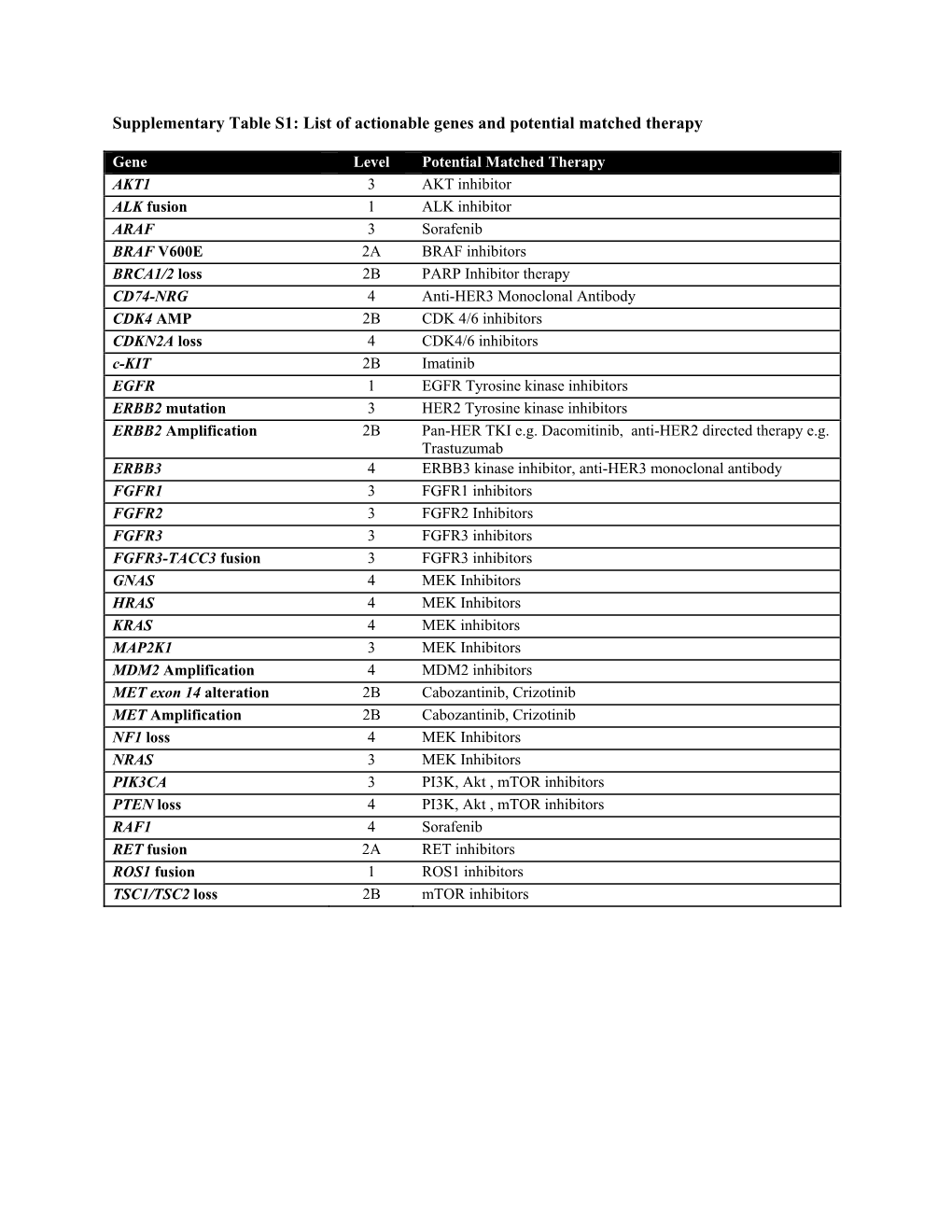
List of Actionable Genes and Potential Matched Therapy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Deregulated Gene Expression Pathways in Myelodysplastic Syndrome Hematopoietic Stem Cells
Leukemia (2010) 24, 756–764 & 2010 Macmillan Publishers Limited All rights reserved 0887-6924/10 $32.00 www.nature.com/leu ORIGINAL ARTICLE Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells A Pellagatti1, M Cazzola2, A Giagounidis3, J Perry1, L Malcovati2, MG Della Porta2,MJa¨dersten4, S Killick5, A Verma6, CJ Norbury7, E Hellstro¨m-Lindberg4, JS Wainscoat1 and J Boultwood1 1LRF Molecular Haematology Unit, NDCLS, John Radcliffe Hospital, Oxford, UK; 2Department of Hematology Oncology, University of Pavia Medical School, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy; 3Medizinische Klinik II, St Johannes Hospital, Duisburg, Germany; 4Division of Hematology, Department of Medicine, Karolinska Institutet, Stockholm, Sweden; 5Department of Haematology, Royal Bournemouth Hospital, Bournemouth, UK; 6Albert Einstein College of Medicine, Bronx, NY, USA and 7Sir William Dunn School of Pathology, University of Oxford, Oxford, UK To gain insight into the molecular pathogenesis of the the World Health Organization.6,7 Patients with refractory myelodysplastic syndromes (MDS), we performed global gene anemia (RA) with or without ringed sideroblasts, according to expression profiling and pathway analysis on the hemato- poietic stem cells (HSC) of 183 MDS patients as compared with the the French–American–British classification, were subdivided HSC of 17 healthy controls. The most significantly deregulated based on the presence or absence of multilineage dysplasia. In pathways in MDS include interferon signaling, thrombopoietin addition, patients with RA with excess blasts (RAEB) were signaling and the Wnt pathways. Among the most signifi- subdivided into two categories, RAEB1 and RAEB2, based on the cantly deregulated gene pathways in early MDS are immuno- percentage of bone marrow blasts. -

Hidden Targets in RAF Signalling Pathways to Block Oncogenic RAS Signalling
G C A T T A C G G C A T genes Review Hidden Targets in RAF Signalling Pathways to Block Oncogenic RAS Signalling Aoife A. Nolan 1, Nourhan K. Aboud 1, Walter Kolch 1,2,* and David Matallanas 1,* 1 Systems Biology Ireland, School of Medicine, University College Dublin, Belfield, Dublin 4, Ireland; [email protected] (A.A.N.); [email protected] (N.K.A.) 2 Conway Institute of Biomolecular & Biomedical Research, University College Dublin, Belfield, Dublin 4, Ireland * Correspondence: [email protected] (W.K.); [email protected] (D.M.) Abstract: Oncogenic RAS (Rat sarcoma) mutations drive more than half of human cancers, and RAS inhibition is the holy grail of oncology. Thirty years of relentless efforts and harsh disappointments have taught us about the intricacies of oncogenic RAS signalling that allow us to now get a pharma- cological grip on this elusive protein. The inhibition of effector pathways, such as the RAF-MEK-ERK pathway, has largely proven disappointing. Thus far, most of these efforts were aimed at blocking the activation of ERK. Here, we discuss RAF-dependent pathways that are regulated through RAF functions independent of catalytic activity and their potential role as targets to block oncogenic RAS signalling. We focus on the now well documented roles of RAF kinase-independent functions in apoptosis, cell cycle progression and cell migration. Keywords: RAF kinase-independent; RAS; MST2; ASK; PLK; RHO-α; apoptosis; cell cycle; cancer therapy Citation: Nolan, A.A.; Aboud, N.K.; Kolch, W.; Matallanas, D. Hidden Targets in RAF Signalling Pathways to Block Oncogenic RAS Signalling. -

RNA-Binding Protein Hnrnpll Regulates Mrna Splicing and Stability During B-Cell to Plasma-Cell Differentiation
RNA-binding protein hnRNPLL regulates mRNA splicing and stability during B-cell to plasma-cell differentiation Xing Changa,b, Bin Lic, and Anjana Raoa,b,d,e,1 Divisions of aSignaling and Gene Expression and cVaccine Discovery, La Jolla Institute for Allergy and Immunology, La Jolla, CA 92037; bSanford Consortium for Regenerative Medicine, La Jolla, CA 92037; and dDepartment of Pharmacology and eMoores Cancer Center, University of California at San Diego, La Jolla, CA 92093 Contributed by Anjana Rao, December 2, 2014 (sent for review July 20, 2014) Posttranscriptional regulation is a major mechanism to rewire the RBP-binding sites, thus validating the specificity of RBP binding transcriptomes during differentiation. Heterogeneous nuclear to coprecipitating RNAs and mapping RBP-binding sites on the RNA-binding protein LL (hnRNPLL) is specifically induced in terminally validated RNAs at close to single-nucleotide resolution (8). differentiated lymphocytes, including effector T cells and plasma Heterogeneous nuclear RNA-binding proteins (hnRNPs) is cells. To study the molecular functions of hnRNPLL at a genome- the term applied to a collection of unrelated nuclear RBPs. wide level, we identified hnRNPLL RNA targets and binding sites in hnRNPLL was identified through a targeted lentiviral shRNA plasma cells through integrated Photoactivatable-Ribonucleoside- screen for regulators of CD45RA to CD45RO switching during Enhanced Cross-Linking and Immunoprecipitation (PAR-CLIP) and memory T-cell development (9) and independently through RNA sequencing. hnRNPLL preferentially recognizes CA dinucleo- two separate screens performed by different groups for exclusion tide-containing sequences in introns and 3′ untranslated regions of CD45 exon 4 in a minigene context (10) and for altered CD44 (UTRs), promotes exon inclusion or exclusion in a context-dependent and CD45R expression on T cells in N-ethyl-N-nitrosourea manner, and stabilizes mRNA when associated with 3′ UTRs. -

Atlas Antibodies in Breast Cancer Research Table of Contents
ATLAS ANTIBODIES IN BREAST CANCER RESEARCH TABLE OF CONTENTS The Human Protein Atlas, Triple A Polyclonals and PrecisA Monoclonals (4-5) Clinical markers (6) Antibodies used in breast cancer research (7-13) Antibodies against MammaPrint and other gene expression test proteins (14-16) Antibodies identified in the Human Protein Atlas (17-14) Finding cancer biomarkers, as exemplified by RBM3, granulin and anillin (19-22) Co-Development program (23) Contact (24) Page 2 (24) Page 3 (24) The Human Protein Atlas: a map of the Human Proteome The Human Protein Atlas (HPA) is a The Human Protein Atlas consortium cell types. All the IHC images for Swedish-based program initiated in is mainly funded by the Knut and Alice the normal tissue have undergone 2003 with the aim to map all the human Wallenberg Foundation. pathology-based annotation of proteins in cells, tissues and organs expression levels. using integration of various omics The Human Protein Atlas consists of technologies, including antibody- six separate parts, each focusing on References based imaging, mass spectrometry- a particular aspect of the genome- 1. Sjöstedt E, et al. (2020) An atlas of the based proteomics, transcriptomics wide analysis of the human proteins: protein-coding genes in the human, pig, and and systems biology. mouse brain. Science 367(6482) 2. Thul PJ, et al. (2017) A subcellular map of • The Tissue Atlas shows the the human proteome. Science. 356(6340): All the data in the knowledge resource distribution of proteins across all eaal3321 is open access to allow scientists both major tissues and organs in the 3. -

ACVR1 Antibody Cat
ACVR1 Antibody Cat. No.: 4791 Western blot analysis of ACVR1 in A549 cell lysate with ACVR1 antibody at 1 μg/mL in (A) the absence and (B) the presence of blocking peptide. Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human, Mouse HOMOLOGY: Predicted species reactivity based on immunogen sequence: Bovine: (100%), Rat: (93%) ACVR1 antibody was raised against a 14 amino acid synthetic peptide near the amino terminus of the human ACVR1. IMMUNOGEN: The immunogen is located within the first 50 amino acids of ACVR1. TESTED APPLICATIONS: ELISA, WB ACVR1 antibody can be used for detection of ACVR1 by Western blot at 1 μg/mL. APPLICATIONS: Antibody validated: Western Blot in human samples. All other applications and species not yet tested. At least four isoforms of ACVR1 are known to exist. This antibody is predicted to have no SPECIFICITY: cross-reactivity to ACVR1B or ACVR1C. POSITIVE CONTROL: 1) Cat. No. 1203 - A549 Cell Lysate Properties October 1, 2021 1 https://www.prosci-inc.com/acvr1-antibody-4791.html PURIFICATION: ACVR1 Antibody is affinity chromatography purified via peptide column. CLONALITY: Polyclonal ISOTYPE: IgG CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: ACVR1 Antibody is supplied in PBS containing 0.02% sodium azide. CONCENTRATION: 1 mg/mL ACVR1 antibody can be stored at 4˚C for three months and -20˚C, stable for up to one STORAGE CONDITIONS: year. As with all antibodies care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: ACVR1 ACVR1 Antibody: FOP, ALK2, SKR1, TSRI, ACTRI, ACVR1A, ACVRLK2, Activin receptor type-1, ALTERNATE NAMES: Activin receptor type I, ACTR-I ACCESSION NO.: NP_001096 PROTEIN GI NO.: 4501895 GENE ID: 90 USER NOTE: Optimal dilutions for each application to be determined by the researcher. -

Funkce CDK12 a CDK13 V Regulaci Transkripce Hana Paculová
MASARYKOVA UNIVERZITA PŘÍRODOVĚDECKÁ FAKULTA ÚSTAV BIOCHEMIE Funkce CDK12 a CDK13 v regulaci transkripce Disertační práce Hana Paculová Školitel: Mgr. Jiří Kohoutek, Ph.D Brno 2018 Bibliogra cký záznam Autorka: Mgr. Hana Paculová Prrodovedecáá aaául,a鏈 Maaarkáova unvverv,a Úa,av bvochemve Název práce: Funáce CDK12 a CDK13 v regulacv ,ranaárvpce Studijní program: Bvochemve Studijní obor: Bvochemve Školitel: Mgr. Jvr Kohou,eá鏈 Ph.D Akademický rok: 2017/2018 Po et stran: 89 Klí ová slova: Ckálvn-dependen,n ávnaaa鏈 CDK12鏈 ,ranaárvpce鏈 RNA polkmeraaa II鏈 raáovvna vaječnáů鏈 CHK1 Bibliographic entry Author: Mgr. Hana Paculová Facul,k oa acvence鏈 Maaarká unvverav,k Department of Biochemistry Title oF dissertation: CDK12 and CDK13 aunc,von vn ,ranacrvp,von regula,von Degree programme: Bvochemva,rk Field oF study: Bvochemva,rk Supervisor: Mgr. Jvr Kohou,eá鏈 Ph.D Academic year: 2017/2018 Number oF pages: 89 Keywords: Ckclvn-dependen ávnaae鏈 CDK12鏈 ,ranacrvp,von鏈 RNA polkmeraae II鏈 ovarvan cancer鏈 CHK1 Abstrakt Ckálvn-dependen,n ávnaaa 12 (CDK12) je ,ranaárvpčn ávnaaa鏈 á,erá rd expreav avých clových genů ,m鏈 že aoaaorkluje RNA polkmeraau II v průbehu elongačn aáe ,ranaárvpce. CDK12 je apojena do neáolváa bunečných preceaů鏈 což ahrnuje odpoveď na pošáoen DNA鏈 vývoj a bunečnou dvaerencvacv a aea,rvh mRNA. CDK12 bkla popaána jaáo jeden genů鏈 á,eré jaou čaa,o mu,ovánk v hvgh-grade aerónm ovarválnm áarcvnomu鏈 nvcméne vlvv ,ech,o mu,ac na aunácv CDK12 a jejvch role v áarcvnogenev dopoaud nebkla a,anovena. Zjva,vlv jame鏈 že ve,švna mu,ac CDK12鏈 á,eré bklk naleenk v nádorech鏈 brán vk,voren áomplexu CDK12 a Ckálvnem K a vnhvbuj ávnaaovou aá,vvv,u CDK12. -

Product Data Sheet
Product Data Sheet ExProfileTM Human AMPK Signaling Related Gene qPCR Array For focused group profiling of human AMPK signaling genes expression Cat. No. QG004-A (4 x 96-well plate, Format A) Cat. No. QG004-B (4 x 96-well plate, Format B) Cat. No. QG004-C (4 x 96-well plate, Format C) Cat. No. QG004-D (4 x 96-well plate, Format D) Cat. No. QG004-E (4 x 96-well plate, Format E) Plates available individually or as a set of 6. Each set contains 336 unique gene primer pairs deposited in one 96-well plate. Introduction The ExProfile human AMPK signaling related gene qPCR array profiles the expression of 336 human genes related to AMPK-mediated signal transduction. These genes are carefully chosen for their close pathway correlation based on a thorough literature search of peer-reviewed publications, mainly including genes that encode AMP-activated protein kinase complex,its regulators and targets involved in many important biological processes, such as glucose uptake, β-oxidation of fatty acids and modulation of insulin secretion. This array allows researchers to study the pathway-related genes to gain understanding of their roles in the different biological processes. QG004 plate 01: 84 unique gene PCR primer pairs QG004 plate 02: 84 unique gene PCR primer pairs QG004 plate 03: 84 unique gene PCR primer pairs QG004 plate 04: 84 unique gene PCR primer pairs Shipping and storage condition Shipped at room temperate Stable for at least 6 months when stored at -20°C Array format GeneCopoeia provides five qPCR array formats (A, B, C, D, and E) suitable for use with the following real- time cyclers. -

The E–Id Protein Axis Modulates the Activities of the PI3K–AKT–Mtorc1
Downloaded from genesdev.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press The E–Id protein axis modulates the activities of the PI3K–AKT–mTORC1– Hif1a and c-myc/p19Arf pathways to suppress innate variant TFH cell development, thymocyte expansion, and lymphomagenesis Masaki Miyazaki,1,8 Kazuko Miyazaki,1,8 Shuwen Chen,1 Vivek Chandra,1 Keisuke Wagatsuma,2 Yasutoshi Agata,2 Hans-Reimer Rodewald,3 Rintaro Saito,4 Aaron N. Chang,5 Nissi Varki,6 Hiroshi Kawamoto,7 and Cornelis Murre1 1Department of Molecular Biology, University of California at San Diego, La Jolla, California 92093, USA; 2Department of Biochemistry and Molecular Biology, Shiga University of Medical School, Shiga 520-2192, Japan; 3Division of Cellular Immunology, German Cancer Research Center, D-69120 Heidelberg, Germany; 4Department of Medicine, University of California at San Diego, La Jolla, California 92093, USA; 5Center for Computational Biology, Institute for Genomic Medicine, University of California at San Diego, La Jolla, California 92093, USA; 6Department of Pathology, University of California at San Diego, La Jolla, California 92093, USA; 7Department of Immunology, Institute for Frontier Medical Sciences, Kyoto University, Kyoto 606-8507, Japan It is now well established that the E and Id protein axis regulates multiple steps in lymphocyte development. However, it remains unknown how E and Id proteins mechanistically enforce and maintain the naı¨ve T-cell fate. Here we show that Id2 and Id3 suppressed the development and expansion of innate variant follicular helper T (TFH) cells. Innate variant TFH cells required major histocompatibility complex (MHC) class I-like signaling and were associated with germinal center B cells. -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -

Epha3 Inhibits Migration and Invasion of Esophageal Cancer Cells by Activating the Mesenchymal‑Epithelial Transition Process
722 INTERNATIONAL JOURNAL OF ONCOLOGY 54: 722-732, 2019 EphA3 inhibits migration and invasion of esophageal cancer cells by activating the mesenchymal‑epithelial transition process XIA CHEN1,2, BIN LU2,3, QIAN MA2, CHENG-DONG JI4 and JIAN-ZHONG LI3 1Key Laboratory, Yangpu Hospital, Tongji University School of Medicine, Shanghai 200090; 2International Joint Cancer Institute; 3Department of Biochemical Pharmacy, Second Military Medical University, Shanghai 200433; 4Department of Scientific Research Management, Yangpu Hospital, Tongji University School of Medicine, Shanghai 200090, P.R. China Received June 13, 2018; Accepted November 2, 2018 DOI: 10.3892/ijo.2018.4639 Abstract. Eph receptor tyrosine kinases are critical for cell-cell Introduction communication during normal and oncogenic development. Eph receptor A3 (EphA3) expression is associated with Esophageal cancer is the eighth most prevalent type of tumor promotion in certain types of cancer; however, it acts cancer worldwide (1,2), of which, esophageal squamous cell as a tumor suppressor in others. The expression levels of carcinoma (ESCC) is a predominant histological type (3). EphA3 and its effects on tumor progression in esophageal Despite advances in diagnostic tools, surgical techniques and squamous cell carcinoma (ESCC) cell lines were determined chemotherapy over the past few decades, the 5-year survival using reverse transcription-quantitative polymerase chain rate for patients with esophageal cancer ranges between reaction analysis and a Transwell invasion assay. The present 15 and 20% (4). Therefore, novel diagnostic tools, therapeutic study demonstrated that EphA3 expression was decreased strategies and molecular prognostic markers are urgently in ESCC tissues and cell lines. Treatment with the DNA required for this disease. -

Pancancer Progression Human Vjune2017
Gene Symbol Accession Alias/Prev Symbol Official Full Name AAMP NM_001087.3 - angio-associated, migratory cell protein ABI3BP NM_015429.3 NESHBP|TARSH ABI family, member 3 (NESH) binding protein ACHE NM_000665.3 ACEE|ARACHE|N-ACHE|YT acetylcholinesterase ACTG2 NM_001615.3 ACT|ACTA3|ACTE|ACTL3|ACTSG actin, gamma 2, smooth muscle, enteric ACVR1 NM_001105.2 ACTRI|ACVR1A|ACVRLK2|ALK2|FOP|SKR1|TSRI activin A receptor, type I ACVR1C NM_145259.2 ACVRLK7|ALK7 activin A receptor, type IC ACVRL1 NM_000020.1 ACVRLK1|ALK-1|ALK1|HHT|HHT2|ORW2|SKR3|TSR-I activin A receptor type II-like 1 ADAM15 NM_207195.1 MDC15 ADAM metallopeptidase domain 15 ADAM17 NM_003183.4 ADAM18|CD156B|CSVP|NISBD|TACE ADAM metallopeptidase domain 17 ADAM28 NM_014265.4 ADAM 28|ADAM23|MDC-L|MDC-Lm|MDC-Ls|MDCL|eMDC II|eMDCII ADAM metallopeptidase domain 28 ADAM8 NM_001109.4 CD156|MS2 ADAM metallopeptidase domain 8 ADAM9 NM_001005845.1 CORD9|MCMP|MDC9|Mltng ADAM metallopeptidase domain 9 ADAMTS1 NM_006988.3 C3-C5|METH1 ADAM metallopeptidase with thrombospondin type 1 motif, 1 ADAMTS12 NM_030955.2 PRO4389 ADAM metallopeptidase with thrombospondin type 1 motif, 12 ADAMTS8 NM_007037.4 ADAM-TS8|METH2 ADAM metallopeptidase with thrombospondin type 1 motif, 8 ADAP1 NM_006869.2 CENTA1|GCS1L|p42IP4 ArfGAP with dual PH domains 1 ADD1 NM_001119.4 ADDA adducin 1 (alpha) ADM2 NM_001253845.1 AM2|dJ579N16.4 adrenomedullin 2 ADRA2B NM_000682.4 ADRA2L1|ADRA2RL1|ADRARL1|ALPHA2BAR|alpha-2BAR adrenoceptor alpha 2B AEBP1 NM_001129.3 ACLP AE binding protein 1 AGGF1 NM_018046.3 GPATC7|GPATCH7|HSU84971|HUS84971|VG5Q -

ACVR1C Antibody Cat
ACVR1C Antibody Cat. No.: 4795 ACVR1C Antibody Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human, Mouse, Rat ACVR1C antibody was raised against a 15 amino acid synthetic peptide near the amino terminus of the human ACVR1C. IMMUNOGEN: The immunogen is located within amino acids 130 - 180 of ACVR1C. TESTED APPLICATIONS: ELISA, WB ACVR1C antibody can be used for detection of ACVR1C by Western blot at 1 and 2 μg/mL. APPLICATIONS: Antibody validated: Western Blot in human samples. All other applications and species not yet tested. SPECIFICITY: This antibody is predicted to have no cross-reactivity to ACVR1 or ACVR1B. POSITIVE CONTROL: 1) Cat. No. 1309 - Human Placenta Tissue Lysate Properties PURIFICATION: ACVR1C Antibody is affinity chromatography purified via peptide column. CLONALITY: Polyclonal September 25, 2021 1 https://www.prosci-inc.com/acvr1c-antibody-4795.html ISOTYPE: IgG CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: ACVR1C Antibody is supplied in PBS containing 0.02% sodium azide. CONCENTRATION: 1 mg/mL ACVR1C antibody can be stored at 4˚C for three months and -20˚C, stable for up to one STORAGE CONDITIONS: year. As with all antibodies care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: ACVR1 ACVR1C Antibody: FOP, ALK2, SKR1, TSRI, ACTRI, ACVR1A, ACVRLK2, Activin receptor ALTERNATE NAMES: type-1, Activin receptor type I, ACTR-I ACCESSION NO.: Q8NER5 PROTEIN GI NO.: 4501895 GENE ID: 90 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References ACVR1C Antibody: Activins are dimeric growth and differentiation factors which belong to the transforming growth factor-beta (TGF-beta) superfamily of structurally related signaling proteins.