Addressing the Problems with Life-Science Databases For
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

HHS Public Access Author Manuscript
HHS Public Access Author manuscript Author Manuscript Author ManuscriptAm J Med Author Manuscript Genet B Neuropsychiatr Author Manuscript Genet. Author manuscript; available in PMC 2015 December 01. Published in final edited form as: Am J Med Genet B Neuropsychiatr Genet. 2014 December ; 0(8): 673–683. doi:10.1002/ajmg.b.32272. Association and ancestry analysis of sequence variants in ADH and ALDH using alcohol-related phenotypes in a Native American community sample Qian Peng1,2,*, Ian R. Gizer3, Ondrej Libiger2, Chris Bizon4, Kirk C. Wilhelmsen4,5, Nicholas J. Schork1, and Cindy L. Ehlers6,* 1 Department of Human Biology, J. Craig Venter Institute, La Jolla, CA 92037 2 Scripps Translational Science Institute, The Scripps Research Institute, La Jolla, CA 92037 3 Department of Psychological Sciences, University of Missouri-Columbia, Columbia, MO 65211 4 Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, NC 27517 5 Department of Genetics and Neurology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 6 Department of Molecular and Cellular Neuroscience, The Scripps Research Institute, La Jolla, CA 92037 Abstract Higher rates of alcohol use and other drug-dependence have been observed in some Native American populations relative to other ethnic groups in the U.S. Previous studies have shown that alcohol dehydrogenase (ADH) genes and aldehyde dehydrogenase (ALDH) genes may affect the risk of development of alcohol dependence, and that polymorphisms within these genes may differentially affect risk for the disorder depending on the ethnic group evaluated. We evaluated variations in the ADH and ALDH genes in a large study investigating risk factors for substance use in a Native American population. -

A Yeast Phenomic Model for the Influence of Warburg Metabolism on Genetic Buffering of Doxorubicin Sean M
Santos and Hartman Cancer & Metabolism (2019) 7:9 https://doi.org/10.1186/s40170-019-0201-3 RESEARCH Open Access A yeast phenomic model for the influence of Warburg metabolism on genetic buffering of doxorubicin Sean M. Santos and John L. Hartman IV* Abstract Background: The influence of the Warburg phenomenon on chemotherapy response is unknown. Saccharomyces cerevisiae mimics the Warburg effect, repressing respiration in the presence of adequate glucose. Yeast phenomic experiments were conducted to assess potential influences of Warburg metabolism on gene-drug interaction underlying the cellular response to doxorubicin. Homologous genes from yeast phenomic and cancer pharmacogenomics data were analyzed to infer evolutionary conservation of gene-drug interaction and predict therapeutic relevance. Methods: Cell proliferation phenotypes (CPPs) of the yeast gene knockout/knockdown library were measured by quantitative high-throughput cell array phenotyping (Q-HTCP), treating with escalating doxorubicin concentrations under conditions of respiratory or glycolytic metabolism. Doxorubicin-gene interaction was quantified by departure of CPPs observed for the doxorubicin-treated mutant strain from that expected based on an interaction model. Recursive expectation-maximization clustering (REMc) and Gene Ontology (GO)-based analyses of interactions identified functional biological modules that differentially buffer or promote doxorubicin cytotoxicity with respect to Warburg metabolism. Yeast phenomic and cancer pharmacogenomics data were integrated to predict differential gene expression causally influencing doxorubicin anti-tumor efficacy. Results: Yeast compromised for genes functioning in chromatin organization, and several other cellular processes are more resistant to doxorubicin under glycolytic conditions. Thus, the Warburg transition appears to alleviate requirements for cellular functions that buffer doxorubicin cytotoxicity in a respiratory context. -

Alcohol Dehydrogenase (ADH1A) Rabbit Polyclonal Antibody Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA323596 Alcohol Dehydrogenase (ADH1A) Rabbit Polyclonal Antibody Product data: Product Type: Primary Antibodies Applications: IHC Recommended Dilution: ELISA: 1:2000-5000, IHC: 1:25-100 Reactivity: Human, Mouse, Rat Host: Rabbit Isotype: IgG Clonality: Polyclonal Immunogen: Fusion protein corresponding to C terminal 300 amino acids of human alcohol dehydrogenase 1A (class I), alpha polypeptide Formulation: PBS pH7.3, 0.05% NaN3, 50% glycerol Concentration: lot specific Purification: Antigen affinity purification Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Gene Name: alcohol dehydrogenase 1A (class I), alpha polypeptide Database Link: NP_000658 Entrez Gene 124 Human P07327 Background: This gene encodes a member of the alcohol dehydrogenase family. The encoded protein is the alpha subunit of class I alcohol dehydrogenase, which consists of several homo- and heterodimers of alpha, beta and gamma subunits. Alcohol dehydrogenases catalyze the oxidation of alcohols to aldehydes. This gene is active in the liver in early fetal life but only weakly active in adult liver. This gene is found in a cluster with six additional alcohol dehydrogenase genes, including those encoding the beta and gamma subunits, on the long arm of chromosome 4. Mutations in this gene may contribute to variation in certain personality traits and substance dependence. Synonyms: ADH1 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. -

Effect of the Allelic Variants of Aldehyde
REVIEW Effect of the allelic variants of aldehyde dehydrogenase ALDH2*2 and alcohol dehydrogenase ADH1B*2 on blood acetaldehyde concentrations Giia-Sheun Peng1 and Shih-Jiun Yin2* 1Department of Neurology, Tri-Service General Hospital, 325 Chenggong Road Section 2, Taipei 114, Taiwan 2Department of Biochemistry, National Defense Medical Center, 161 Minchuan East Road Section 6, Taipei 114, Taiwan *Correspondence to: Tel: þ886 2 8792 3100 ext 18800; Fax: þ886 2 8792 4818; E-mail: [email protected] Date received (in revised form): 30th July 2008 Abstract Alcoholism is a complex behavioural disorder. Molecular genetics studies have identified numerous candidate genes associated with alcoholism. It is crucial to verify the disease susceptibility genes by correlating the pinpointed allelic variations to the causal phenotypes. Alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) are the principal enzymes responsible for ethanol metabolism in humans. Both ADH and ALDH exhibit functional polymorphisms among racial populations; these polymorphisms have been shown to be the important genetic determinants in ethanol metabolism and alcoholism. Here, we briefly review recent advances in genomic studies of human ADH/ALDH families and alcoholism, with an emphasis on the pharmacogenetic consequences of venous blood acetaldehyde in the different ALDH2 genotypes following the intake of various doses of ethanol. This paper illustrates a paradigmatic example of phenotypic verifications in a protective disease gene for substance abuse. Keywords: alcohol dehydrogenase, aldehyde dehydrogenase, single nucleotide polymorphism, alcoholism, ethanol metab- olism, blood acetaldehyde Introduction Alcohol dehydrogenase (ADH) and aldehyde dehy- exposure and the concentrations of ethanol and its drogenase (ALDH) are the principal enzymes respon- metabolite acetaldehyde attained in body fluids and sible for hepatic metabolism of ethanol.1 Human tissue within that period. -

The MDR Superfamily
Cell. Mol. Life Sci. 65 (2008) 3879 – 3894 View metadata,1420-682X/08/243879-16 citation and similar papers at core.ac.uk Cellular and Molecular Life Sciencesbrought to you by CORE DOI 10.1007/s00018-008-8587-z provided by Springer - Publisher Connector Birkhuser Verlag, Basel, 2008 The MDR superfamily B. Perssona,b,*, J. Hedlunda and H. Jçrnvallc a IFM Bioinformatics, Linkçping University, 581 83 Linkçping (Sweden), Fax: +4613137568, e-mail: [email protected] b Dept of Cell and Molecular Biology, Karolinska Institutet, 171 77 Stockholm (Sweden) c Dept of Medical Biochemistry and Biophysics, Karolinska Institutet, 171 77 Stockholm (Sweden) Online First 14 November 2008 Abstract. The MDR superfamily with ~350-residue ments. Subsequent recognitions now define at least 40 subunits contains the classical liver alcohol dehydro- human MDR members in the Uniprot database genase (ADH), quinone reductase, leukotriene B4 (correspondingACHTUNGRE to 25 genes when excluding close dehydrogenase and many more forms. ADH is a homologues), and in all species at least 10888 entries. dimeric zinc metalloprotein and occurs as five differ- Overall, variability is large, but like for many dehy- ent classes in humans, resulting from gene duplica- drogenases, subdivided into constant and variable tions during vertebrate evolution, the first one traced forms, corresponding to household and emerging to ~500 MYA (million years ago) from an ancestral enzyme activities, respectively. This review covers formaldehyde dehydrogenase line. Like many dupli- basic facts and describes eight large MDR families and cations at that time, it correlates with enzymogenesis nine smaller families. Combined, they have specific of new activities, contributing to conditions for substrates in metabolic pathways, some with wide emergence of vertebrate land life from osseous fish. -

Alcohol Dehydrogenase (ADH1A) (NM 000667) Human Recombinant Protein Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TP315367 Alcohol Dehydrogenase (ADH1A) (NM_000667) Human Recombinant Protein Product data: Product Type: Recombinant Proteins Description: Recombinant protein of human alcohol dehydrogenase 1A (class I), alpha polypeptide (ADH1A) Species: Human Expression Host: HEK293T Tag: C-Myc/DDK Predicted MW: 39.7 kDa Concentration: >50 ug/mL as determined by microplate BCA method Purity: > 80% as determined by SDS-PAGE and Coomassie blue staining Buffer: 25 mM Tris.HCl, pH 7.3, 100 mM glycine, 10% glycerol Preparation: Recombinant protein was captured through anti-DDK affinity column followed by conventional chromatography steps. Storage: Store at -80°C. Stability: Stable for 12 months from the date of receipt of the product under proper storage and handling conditions. Avoid repeated freeze-thaw cycles. RefSeq: NP_000658 Locus ID: 124 UniProt ID: P07327 RefSeq Size: 1456 Cytogenetics: 4q23 RefSeq ORF: 1125 Synonyms: ADH1 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 Alcohol Dehydrogenase (ADH1A) (NM_000667) Human Recombinant Protein – TP315367 Summary: This gene encodes a member of the alcohol dehydrogenase family. The encoded protein is the alpha subunit of class I alcohol dehydrogenase, which consists of several homo- and heterodimers of alpha, beta and gamma subunits. Alcohol dehydrogenases catalyze the oxidation of alcohols to aldehydes. This gene is active in the liver in early fetal life but only weakly active in adult liver. -

Discovering Pharmacogenomic Variants and Pathways with the Epigenome and Spatial Genome
The Pharmacoepigenomics Informatics Pipeline and H-GREEN Hi-C Compiler: Discovering Pharmacogenomic Variants and Pathways with the Epigenome and Spatial Genome by Ari Lawrence Allyn-Feuer A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Bioinformatics) in the University of Michigan 2018 Doctoral Committee: Professor Brian D. Athey, Chair Assistant Professor Alan P. Boyle Professor Ivo D. Dinov Research Professor Gerald A. Higgins Professor Wolfgang Sadee, The Ohio State University Ari Lawrence Allyn-Feuer [email protected] ORCID iD: 0000-0002-8379-2765 © Ari Allyn-Feuer 2018 Dedication In the epilogue of Altneuland, Theodor Herzl famously wrote: “Dreams are not so different from deeds as some may think. All the deeds of men are dreams at first, and become dreams in the end.” Medical advances undergo a similar progression, from invisible to visible and back. Before they are accomplished, advances in the physician’s art are illegible: no one differentiates from the rest the suffering and death which could be alleviated with methods which do not yet exist. Unavoidable ills have none of the moral force of avoidable ones. Then, for a brief period, beginning shortly before it is deployed in mainstream practice, and slowly concluding over the generation after it becomes widespread, an advance is visible. People see the improvements and celebrate them. Then, subsequently, for the rest of history, if we are lucky, such an advance is more invisible than it was before it was invented. No one tallies the children who do not get polio, the firm ground that used to be a malarial swamp, or the quiet fact of sanitation. -

Alcohol Dehydrogenase Antibody / ADH (R32497)
Alcohol dehydrogenase Antibody / ADH (R32497) Catalog No. Formulation Size R32497 0.5mg/ml if reconstituted with 0.2ml sterile DI water 100 ug Bulk quote request Availability 1-3 business days Species Reactivity Human, Mouse, Rat Format Antigen affinity purified Clonality Polyclonal (rabbit origin) Isotype Rabbit IgG Purity Antigen affinity Buffer Lyophilized from 1X PBS with 2.5% BSA and 0.025% sodium azide UniProt P07327 Localization Cytoplasmic Applications Western blot : 0.5-1ug/ml Immunohistochemistry (FFPE) : 1-2ug/ml Immunofluorescence (FFPE) : 2-4ug/ml Flow cytometry : 1-3ug/million cells Limitations This Alcohol dehydrogenase antibody is available for research use only. Immunofluorescent staining of FFPE human U-2 OS cells with Alcohol dehydrogenase antibody (green) and DAPI nuclear stain (blue). HIER: steam section in pH6 citrate buffer for 20 min. IHC testing of FFPE human liver cancer tissue with Alcohol dehydrogenase antibody at 1ug/ml. HIER: steam sections in pH6 citrate buffer for 20 min. Western blot testing of 1) rat lung, 2) mouse liver and 3) human HepG2 lysate with Alcohol dehydrogenase antibody at 0.5ug/ml. Predicted molecular weight ~40 kDa. Flow cytometry testing of human U-2 OS cells with Alcohol dehydrogenase antibody at 1ug/million cells (blocked with goat sera); Red=cells alone, Green=isotype control, Blue= Alcohol dehydrogenase antibody. Description Alcohol dehydrogenase 1A is an enzyme that in humans is encoded by the ADH1A gene. This gene encodes a member of the alcohol dehydrogenase family. The encoded protein is the alpha subunit of class I alcohol dehydrogenase, which consists of several homo- and heterodimers of alpha, beta and gamma subunits. -

(12) United States Patent (10) Patent No.: US 9.273,330 B2 Bramucci Et Al
US00927.333 OB2 (12) United States Patent (10) Patent No.: US 9.273,330 B2 Bramucci et al. (45) Date of Patent: Mar. 1, 2016 (54) BUTANOL TOLERANCE IN 8,206,970 B2 6/2012 Eliot et al. MCROORGANISMS 8,222,017 B2 7/2012 Li et al. 8,241,878 B2 8/2012 Anthony et al. (71) Applicant: Butamax Advanced Biofuels LLC, 3. E: 858: Ration s al Wilmington, DE (US) 8,372,612 B2 2/2013 Larossa et al. 8,389,252 B2 3/2013 Larossa (72) Inventors: Michael G. Bramucci, Boothwyn, PA 8,455,224 B2 6/2013 Paul (US); Vasantha Nagarajan, 8,455.225 B2 6/2013 Bramucci et al. Wilmington,s DE (US) - E.Kw E. E. E.a 8,557.562 B2 10/2013 Bramucci et al. (73) Assignee: Butamax Advanced Biofuels LLC, 8,614,085 B2 12/2013 Van Dyk Wilmington, DE (US) 8,637,281 B2 1/2014 Paul et al. 8,637,289 B2 1/2014 Anthony et al. (*)c Notice:- r Sibi tO E. site th still 8,669,0948,652,823 B2 3/20142/2014 AnthonyFlint et al. et al. patent 1s extended or adjusted under 8,691,540 B2 4/2014 Bramucci et al. U.S.C. 154(b) by 2 days. 8,735,114 B2 5/2014 Donaldson et al. 8,765,433 B2 7/2014 Satagopan et al. (21) Appl. No.: 14/045,506 8,785,166 B2 7/2014 Anthony 8,795,992 B2 8/2014 Bramucci et al. (22) Filed: Oct. 3, 2013 5%. -

The Xenopus Alcohol Dehydrogenase Gene Family
Borràs et al. BMC Genomics 2014, 15:216 http://www.biomedcentral.com/1471-2164/15/216 RESEARCH ARTICLE Open Access The Xenopus alcohol dehydrogenase gene family: characterization and comparative analysis incorporating amphibian and reptilian genomes Emma Borràs1, Ricard Albalat2, Gregg Duester3, Xavier Parés1 and Jaume Farrés1* Abstract Background: The alcohol dehydrogenase (ADH) gene family uniquely illustrates the concept of enzymogenesis. In vertebrates, tandem duplications gave rise to a multiplicity of forms that have been classified in eight enzyme classes, according to primary structure and function. Some of these classes appear to be exclusive of particular organisms, such as the frog ADH8, a unique NADP+-dependent ADH enzyme. This work describes the ADH system of Xenopus, as a model organism, and explores the first amphibian and reptilian genomes released in order to contribute towards a better knowledge of the vertebrate ADH gene family. Results: Xenopus cDNA and genomic sequences along with expressed sequence tags (ESTs) were used in phylogenetic analyses and structure-function correlations of amphibian ADHs. Novel ADH sequences identified in the genomes of Anolis carolinensis (anole lizard) and Pelodiscus sinensis (turtle) were also included in these studies. Tissue and stage-specific libraries provided expression data, which has been supported by mRNA detection in Xenopus laevis tissues and regulatory elements in promoter regions. Exon-intron boundaries, position and orientation of ADH genes were deduced from the amphibian and reptilian genome assemblies, thus revealing syntenic regions and gene rearrangements with respect to the human genome. Our results reveal the high complexity of the ADH system in amphibians, with eleven genes, coding for seven enzyme classes in Xenopus tropicalis. -

Use of Network Pharmacology to Investigate the Mechanism of the Compound Xuanju Capsule in the Treatment of Rheumatoid Arthritis
Hindawi BioMed Research International Volume 2021, Article ID 5568791, 14 pages https://doi.org/10.1155/2021/5568791 Research Article Use of Network Pharmacology to Investigate the Mechanism of the Compound Xuanju Capsule in the Treatment of Rheumatoid Arthritis Wenyang Wei, Wanpeng Lu, Xiaolong Chen, Yunfeng Yang, and Mengkai Zheng Academic Research and Development Center of Zhejiang Strong Pharmaceutical Co., Ltd., Hangzhou, 310053 Zhejiang, China Correspondence should be addressed to Mengkai Zheng; [email protected] Received 24 February 2021; Accepted 24 July 2021; Published 10 August 2021 Academic Editor: Nadeem Sheikh Copyright © 2021 Wenyang Wei et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Objective. To clarify the therapeutic mechanisms of compound Xuanju capsule-treated rheumatoid arthritis (RA) based on network pharmacology tactics. Method. The TCMSP, TCMID and STITCH databases were used to screen the active ingredients and targets in the compound Xuanju capsule; the OMIM, TTD, PharmGKB and GeneCards databases were applied to screen the RA-related disease targets. Then, the obtained targets were imported into Cytoscape 3.7.1 software to construct the active ingredient-target network and the RA-related disease-target network. The active ingredient-target PPI network, the RA-related disease-target PPI network and the common target PPI network were built by using the STRING platform and Cytoscape 3.7.1 software. The GO and KEGG analyses of the common targets were analyzed by using the Metascape and Bioinformatics online tools. -
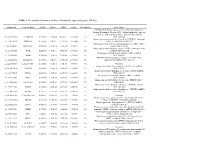
Table 1 the Statistical Metrics for Key Differentially Expressed Genes (Degs)
Table 1 The statistical metrics for key differentially expressed genes (DEGs) Agiliant Id Gene Symbol logFC pValue FDR tvalue Regulation Gene Name PREDICTED: Homo sapiens similar to Filamin-C (Gamma- filamin) (Filamin-2) (Protein FLNc) (Actin-binding-like protein) (ABP-L) (ABP-280-like protein) (LOC649056), mRNA A_24_P237896 LOC649056 0.509843 9.18E-14 4.54E-11 12.07302 Up [XR_018580] Homo sapiens programmed cell death 10 (PDCD10), transcript A_23_P18325 PDCD10 0.243111 2.8E-12 9.24E-10 10.62808 Up variant 1, mRNA [NM_007217] Homo sapiens Rho GTPase activating protein 27 (ARHGAP27), A_23_P141335 ARHGAP27 0.492709 3.97E-12 1.22E-09 10.48571 Up mRNA [NM_199282] Homo sapiens tubby homolog (mouse) (TUB), transcript variant A_23_P53110 TUB 0.528219 1.77E-11 4.56E-09 9.891033 Up 1, mRNA [NM_003320] Homo sapiens MyoD family inhibitor (MDFI), mRNA A_23_P42168 MDFI 0.314474 1.81E-10 3.74E-08 8.998697 Up [NM_005586] PREDICTED: Homo sapiens hypothetical LOC644701 A_32_P56890 LOC644701 0.444703 3.6E-10 7.09E-08 8.743973 Up (LOC644701), mRNA [XM_932316] A_32_P167111 A_32_P167111 0.873588 7.41E-10 1.4E-07 8.47781 Up Unknown Homo sapiens zinc finger protein 784 (ZNF784), mRNA A_24_P221424 ZNF784 0.686781 9.18E-10 1.68E-07 8.399687 Up [NM_203374] Homo sapiens lin-28 homolog (C. elegans) (LIN28), mRNA A_23_P74895 LIN28 0.218876 1.27E-09 2.24E-07 8.282224 Up [NM_024674] Homo sapiens ribosomal protein L5 (RPL5), mRNA A_23_P12140 RPL5 0.247598 1.81E-09 3.11E-07 8.154317 Up [NM_000969] Homo sapiens cDNA FLJ43841 fis, clone TESTI4006137.