Evolution of Host Range in the Follicle Mite Demodex Kutzeri
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

George Et Al 1992 Louse Mite Infestations Domestic Animals Nigeria
Trop. Anita. Hlth Prod. (1992) 24, 121-124 LOUSE AND MITE INFESTATION IN DOMESTIC ANIMALS IN NORTHERN NIGERIA J. B. D. GEORGE, S. OTOBO, J. OGUNLEYEand B. ADEDIMINIYI Department of Veterinary Parasitology and Entomology, Faculty of Veterinary Medicine, Ahmadu Bello University, Zaria, Nigeria SUMMARY Records of domestic animals brought to the Veterinary Entomology Laboratory for diagnosis of suspected lice and mite infestation over a 10 year period were analysed. From a total of 794 suspected cases, 137 (17.3%) and247 (31.1%) were positive for lice and mange mites respectively. The most common lice species recorded were Linognathus vituli (66.7%) on cattle, L. ovillus (83.3%) on sheep, Haematopinus suis (100%) on pigs and Menacanthus stramineus (54.5%) on poultry. Other lice species recorded included Haematopinus bovis and Solenopotes capillatus on cattle, Damalinia ovis on sheep, Linognathus stenopsis and Mena- canthus stramineus on goats, Goniocotes sp. on a horse, Linognathus setosus and Menacanthus stramineus on dogs, Goniodes gigas, Lipeurus caponis, Menopon gallinae and Chelopistes meleagrides on poultry. The most common mite species were Demodex folliculorum on cattle (96.9%) and on dogs (80.8%), Sarcoptes scabiei on pigs (100%) and Notoedres cati (80.3%) on rabbits. Other mite species included Psoroptes communis, Cheyletiella parasitivorax, Ornithonyssus gallinae and Dermanyssus gallinae. INTRODUCTION Lice and mite infestations often cause stress and loss of condition (Schillhorn van Veen and Mohammed, 1975; Bamidele and Amakiri, 1978; Idowu and Adetunji, 1981; Okon, 1981). Usually a dermatitis is manifested which is characterised by alopecia and necrotic foci. There is also intense pruritus (especially with mange) which leads to biting and vigorous scratching of affected parts (Lapage, 1968; Sweatman, 1973; Idowu and Adetunji, 1981). -

Demodex Folliculorum in Nasal Discharge: a Case Report of Yet Unknown Significance
Global Journal of Otolaryngology ISSN 2474-7556 Case Report Glob J Otolaryngol Volume 18 Issue 2 - November 2018 Copyright © All rights are reserved by Neelam Riyaz Attar DOI: 10.19080/GJO.2018.18.555984 Demodex Folliculorum In Nasal Discharge: A Case Report of Yet Unknown Significance Neelam R Attar* Department of Microbiology, Assistant Professor, India Submission: November 15, 2018; Published: November 26, 2018 *Corresponding author: Neelam Riyaz Attar, Department of Microbiology, Assistant Professor, Niyaz Manzil, Kolhapur, India Abstract Demodex mites are the parasites residing in the pilo-sebaceous follicle and sebaceous gland. They are frequently isolated from cases of Demodexfolliculitis, folliculorum. Rosacea and various other inflammatory dermatoses. We report a possible case of demodecosis in patient of mucormycosis. Nasal scraping and discharge was negative for fungal elements but contained high density of gravid Demodex mites. The species was identified as Keywords: Demodex mites; Demodecosis; Nasal scrapping; Diabetes mellitus Introduction on SDA agar and incubated at 37C and 25C for 4 weeks. The Demodex mites are the normal inhabitants of pilosebaceous culture was reported as negative. Diagnosis of mucormycosis unit and gland. Demodex folliculorum and Demodex brevis are the two species found all over the body especially areas dense presence of broad non septate hyphae with right angle branching. in sebaceous glands including face, neck, back and chest [1- was confirmed by biopsy of right middle meatus which showed 3]. Previously thought to be harmless commensal they are now as it was longer than its counterpart D. brevis (Size 0.1 – 0.2 recently implicated as the causative agent of many dermatoses Demodex species was identified as that of Demodex folliculorum mm) [6]. -

Deconstructing Canine Demodicosis”
TESIS DOCTORAL TITULO: “Deconstructing canine demodicosis” AUTOR: Ivan Ravera DIRECTORES: Lluís Ferrer, Mar Bardagí, Laia Solano Gallego. PROGRAMA DE DOCTORADO: Medicina i Sanitat Animals DEPARTAMENTO: Medicina i Cirurgia Animals UNIVERSIDAD: Universitat Autònoma de Barcelona 2015 Dr. Lluis Ferrer i Caubet, Dra. Mar Bardagí i Ametlla y Dra. Laia María Solano Gallego, docentes del Departamento de Medicina y Cirugía Animales de la Universidad Autónoma de Barcelona, HACEN CONSTAR: Que la memoria titulada “Deconstructing canine demodicosis” presentada por el licenciado Ivan Ravera para optar al título de Doctor por la Universidad Autónoma de Barcelona, se ha realizado bajo nuestra dirección, y considerada terminada, autorizo su presentación para que pueda ser juzgada por el tribunal correspondiente. Y por tanto, para que conste firmo el presente escrito. Bellaterra, el 23 de Septiembre de 2015. Dr. Lluis Ferrer, Dra. Mar Bardagi, Ivan Ravera Dra. Laia Solano Gallego Directores de la tesis doctoral Doctorando AGRADECIMIENTOS A los alquimistas de guantes azules A los otros luchadores - Ester Blasco - Diana Ferreira - Lola Pérez - Isabel Casanova - Aida Neira - Gina Doria - Blanca Pérez - Marc Isidoro - Mercedes Márquez - Llorenç Grau - Anna Domènech - los internos del HCV-UAB - Elena García - los residentes del HCV-UAB - Neus Ferrer - Manuela Costa A los veterinarios - Sergio Villanueva - del HCV-UAB - Marta Carbonell - dermatólogos españoles - Mónica Roldán - Centre d’Atenció d’Animals de Companyia del Maresme A los sensacionales genetistas -

Arthropods of Public Health Significance in California
ARTHROPODS OF PUBLIC HEALTH SIGNIFICANCE IN CALIFORNIA California Department of Public Health Vector Control Technician Certification Training Manual Category C ARTHROPODS OF PUBLIC HEALTH SIGNIFICANCE IN CALIFORNIA Category C: Arthropods A Training Manual for Vector Control Technician’s Certification Examination Administered by the California Department of Health Services Edited by Richard P. Meyer, Ph.D. and Minoo B. Madon M V C A s s o c i a t i o n of C a l i f o r n i a MOSQUITO and VECTOR CONTROL ASSOCIATION of CALIFORNIA 660 J Street, Suite 480, Sacramento, CA 95814 Date of Publication - 2002 This is a publication of the MOSQUITO and VECTOR CONTROL ASSOCIATION of CALIFORNIA For other MVCAC publications or further informaiton, contact: MVCAC 660 J Street, Suite 480 Sacramento, CA 95814 Telephone: (916) 440-0826 Fax: (916) 442-4182 E-Mail: [email protected] Web Site: http://www.mvcac.org Copyright © MVCAC 2002. All rights reserved. ii Arthropods of Public Health Significance CONTENTS PREFACE ........................................................................................................................................ v DIRECTORY OF CONTRIBUTORS.............................................................................................. vii 1 EPIDEMIOLOGY OF VECTOR-BORNE DISEASES ..................................... Bruce F. Eldridge 1 2 FUNDAMENTALS OF ENTOMOLOGY.......................................................... Richard P. Meyer 11 3 COCKROACHES ........................................................................................... -

Role of Demodex Infestation in Blepharitis and Coconut Oil As a Treatment Option
Indian Journal of Clinical and Experimental Ophthalmology 2020;6(2):270–275 Content available at: iponlinejournal.com Indian Journal of Clinical and Experimental Ophthalmology Journal homepage: www.innovativepublication.com Original Research Article Role of demodex infestation in blepharitis and coconut oil as a treatment option Suresha A R1, Sadhwini M H1,* 1Dept. of Ophthalmology, JJM Medical College, Davangere, Karnataka, India ARTICLEINFO ABSTRACT Article history: Purpose: To assess incidence of demodex species, correlate ocular symptomatology, evaluate efficacy of Received 03-01-2020 coconut oil as treatment method in all types of blepharitis. Accepted 06-02-2020 Materials and Methods: 30 patients with anterior & mixed blepharitis, meibomian gland dysfunction Available online 16-06-2020 & non-specific irritation were enrolled for study. History taken & examined clinically. 2 lashes/lid were sampled & mounted on slides with normal saline & observed under light microscope. Number of mites counted. Patients positive for demodex were treated with coconut oil application over lid margins & Keywords: reviewed after 3 weeks. Anterior blepharitis Results: Incidence of demodex was 40% & it increased with age. Demodex was commonly associated with Demodex infestation meibomian gland dysfunction, non-specific irritation, madarosis, cloudy & toothpaste like meibum quality. Meibomian gland dysfunction Burning sensation and itching were common complaints. At 3rd week, all patients were symptom-free. Non-specific irritation Mite count dropped by 52.8% but were not eliminated. Conclusion: Demodex infestation is often overlooked but it is associated with about half of blepharitis cases. Hence further evaluation should be considered. Coconut oil is an easily available mode of treatment & helps reduce symptoms and mite counts. © 2020 Published by Innovative Publication. -

Parasitology JWST138-Fm JWST138-Gunn February 21, 2012 16:59 Printer Name: Yet to Come P1: OTA/XYZ P2: ABC
JWST138-fm JWST138-Gunn February 21, 2012 16:59 Printer Name: Yet to Come P1: OTA/XYZ P2: ABC Parasitology JWST138-fm JWST138-Gunn February 21, 2012 16:59 Printer Name: Yet to Come P1: OTA/XYZ P2: ABC Parasitology An Integrated Approach Alan Gunn Liverpool John Moores University, Liverpool, UK Sarah J. Pitt University of Brighton, UK Brighton and Sussex University Hospitals NHS Trust, Brighton, UK A John Wiley & Sons, Ltd., Publication JWST138-fm JWST138-Gunn February 21, 2012 16:59 Printer Name: Yet to Come P1: OTA/XYZ P2: ABC This edition first published 2012 © 2012 by by John Wiley & Sons, Ltd Wiley-Blackwell is an imprint of John Wiley & Sons, formed by the merger of Wiley’s global Scientific, Technical and Medical business with Blackwell Publishing. Registered Office John Wiley & Sons Ltd, The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, UK Editorial Offices 9600 Garsington Road, Oxford, OX4 2DQ, UK The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, UK 111 River Street, Hoboken, NJ 07030-5774, USA For details of our global editorial offices, for customer services and for information about how to apply for permission to reuse the copyright material in this book please see our website at www.wiley.com/wiley-blackwell. The right of the author to be identified as the author of this work has been asserted in accordance with the UK Copyright, Designs and Patents Act 1988. All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, except as permitted by the UK Copyright, Designs and Patents Act 1988, without the prior permission of the publisher. -

Acne Vulgaris, Rosacea, Seborrheic Dermatitisଝ,ଝଝ
An Bras Dermatol. 2020;95(2):187---193 Anais Brasileiros de Dermatologia www.anaisdedermatologia.org.br INVESTIGATION Demodex folliculorum infestations in common facial dermatoses: acne vulgaris, rosacea, seborrheic dermatitisଝ,ଝଝ ∗ Ezgi Aktas¸ Karabay , Aslı Aksu C¸erman Department of Dermatology and Venereology, Faculty of Medicine, Bahc¸es¸ehir University, Istanbul, Turkey Received 18 March 2019; accepted 26 August 2019 Available online 12 February 2020 Abstract KEYWORDS Background: Demodex mites are found on the skin of many healthy individuals. Demodex mites Acne vulgaris; in high densities are considered to play a pathogenic role. Dermatitis, Objective: To investigate the association between Demodex infestation and the three most seborrheic; common facial dermatoses: acne vulgaris, rosacea and seborrheic dermatitis. Rosacea Methods: This prospective, observational case-control study included 127 patients (43 with acne vulgaris, 43 with rosacea and 41 with seborrheic dermatitis) and 77 healthy controls. The presence of demodicosis was evaluated by standardized skin surface biopsy in both the patient and control groups. Results: In terms of gender and age, no significant difference was found between the patients and controls (p > 0.05). Demodex infestation rates were significantly higher in patients than in controls (p = 0.001). Demodex infestation rates were significantly higher in the rosacea group than acne vulgaris and seborrheic dermatitis groups and controls (p = 0.001; p = 0.024; p = 0.001, respectively). Demodex infestation was found to be significantly higher in the acne vulgaris and seborrheic dermatitis groups than in controls (p = 0.001 and p = 0.001, respectively). No difference was observed between the acne vulgaris and seborrheic dermatitis groups in terms of demodicosis (p = 0.294). -

Demodicidosis of the Nipple
186 Letters to the Editor Demodicidosis of the Nipple Thomas Jansen1, Falk Georges Bechara2, Markus Stu¨cker2 and Peter Altmeyer2 Department of Dermatology, 1University of Essen, Hufelandstrasse 55, DE-45122 Essen, and 2Ruhr-University Bochum, Germany E-mail: [email protected] Accepted July 19, 2004. Sir, 3 weeks. At the end of treatment, no Demodex mites Demodex mites are common ectoparasites of human could be demonstrated by skin surface biopsy (cyano- skin (1). Two species, Demodex folliculorum and the acrylate technique) from the nipple. No recurrences smaller D. brevis, inhabit the human pilosebaceous unit have been observed in a 12-month follow-up. and are found in individuals of all ages except neonates. They inhabit almost every area of human skin but show a predilection for the face, especially the nose and nasolabial folds. Cases in which demodicidosis devel- oped on other sites than the face were associated with the acquired immunodeficiency syndrome (AIDS) (2), diabetes mellitus, chronic liver disease (3), or mycosis fungoides (4). This is a report of an otherwise healthy patient affected with unilateral eczema-like demodici- dosis of the nipple. CASE REPORT A 53-year-old Caucasian man presented with pruritic skin lesions on the right nipple that had been present for 8 months. He had not used topical or systemic corticosteroids or other immunosuppressive drugs. Otherwise he was in good health. The personal and familial history of atopy was negative. Clinical examination showed mild erythema with scaling on the right nipple. The remainder of the skin surface was normal. Histopathology of a skin biopsy revealed a dense perifollicular lymphocytic infiltrate (Fig. -

Studies on the Distribution and Habitat of Demodex Folliculorum
Portland State University PDXScholar Dissertations and Theses Dissertations and Theses 5-1-1970 Studies on the distribution and habitat of Demodex folliculorum Michael James Gimbol Portland State University Follow this and additional works at: https://pdxscholar.library.pdx.edu/open_access_etds Let us know how access to this document benefits ou.y Recommended Citation Gimbol, Michael James, "Studies on the distribution and habitat of Demodex folliculorum" (1970). Dissertations and Theses. Paper 58. https://doi.org/10.15760/etd.58 This Thesis is brought to you for free and open access. It has been accepted for inclusion in Dissertations and Theses by an authorized administrator of PDXScholar. Please contact us if we can make this document more accessible: [email protected]. AN ABSTRACT OF TILE THESIS OF Michael Jam·es Gimbol for the Master of Science in Biology presented May 2Z, 1970. Ti t If3 ~ Stttd i es on the Di s t r ibt1. ti on .and Hah i tat of De.!"!,:..oc0!.?i foll iculor-tur.. • ..........-..-.~ .......------~ APPROVED BY ~ilEl\;ffiERS OF THE; THES IS C01VIJHl TTE.E;: :.o;....-.,.,~""tJl ---.............. ~··_~ ..····~ . ..__ Richard B~ Forbes ="9 - , The present study was undertalcen to determine the Derr.:..c:~l ~ 9~:~!_~Tl1~. inci dence of i nfestation \vi til € f 01 i j a s lei. n mite parasitizing man, and to investi.gate those factors' which influence its occurrence.. These factors were the personal hygiene, sex, age a.n.d. skin conditIon ()f the human host~ A detailed view of the habitat of this parasite is also included so that the controversy 8urrotu"1ding its I'ole as a possible pathogen or vector of disease could be explored in 1i ght of the current }(11,O\;v!ed ge o(l A ntnnber of si tes of infestat.ion were sampled by a 2 number of different techniques. -

Complete Mitochondrial Genomes of the Human Follicle Mites Demodex Brevis and D
Palopoli et al. BMC Genomics 2014, 15:1124 http://www.biomedcentral.com/1471-2164/15/1124 RESEARCH ARTICLE Open Access Complete mitochondrial genomes of the human follicle mites Demodex brevis and D. folliculorum: novel gene arrangement, truncated tRNA genes, and ancient divergence between species Michael F Palopoli*, Samuel Minot, Dorothy Pei, Alicia Satterly and Julie Endrizzi Abstract Background: Follicle mites of the genus Demodex are found on a wide diversity of mammals, including humans; surprisingly little is known, however, about the evolution of this association. Additional sequence information promises to facilitate studies of Demodex variation within and between host species. Here we report the complete mitochondrial genome sequences of two species of Demodex known to live on humans—Demodex brevis and D. folliculorum—which are the first such genomes available for any member of the genus. We analyzed these sequences to gain insight into the evolution of mitochondrial genomes within the Acariformes. We also used relaxed molecular clock analyses, based on alignments of mitochondrial proteins, to estimate the time of divergence between these two species. Results: Both Demodex genomes shared a novel gene order that differs substantially from the ancestral chelicerate pattern, with transfer RNA (tRNA) genes apparently having moved much more often than other genes. Mitochondrial tRNA genes of both species were unusually short, with most of them unable to encode tRNAs that could fold into the canonical cloverleaf structure; indeed, several examples lacked both D- and T-arms. Finally, the high level of sequence divergence observed between these species suggests that these two lineages last shared a common ancestor no more recently than about 87 mya. -
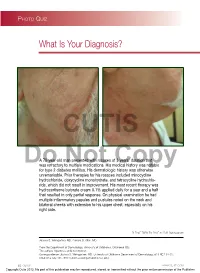
Demodex Folliculitis
Photo Quiz What Is Your Diagnosis? CUTIS A 79-year-old man presented with rosacea of 3 years’ duration that Dowas refractory toNot multiple medications. His medicalCopy history was notable for type 2 diabetes mellitus. His dermatologic history was otherwise unremarkable. Prior therapies for his rosacea included minocycline hydrochloride, doxycycline monohydrate, and tetracycline hydrochlo- ride, which did not result in improvement. His most recent therapy was hydrocortisone butyrate cream 0.1% applied daily for a year and a half that resulted in only partial response. On physical examination he had multiple inflammatory papules and pustules noted on the neck and bilateral cheeks with extension to his upper chest, especially on his right side. PLEASE TURN TO PAGE 65 FOR DISCUSSION Joshua S. Weingartner, MD; Pamela S. Allen, MD From the Department of Dermatology, University of Oklahoma, Oklahoma City. The authors report no conflict of interest. Correspondence: Joshua S. Weingartner, MD, University of Oklahoma Department of Dermatology, 619 NE 13th St, Oklahoma City, OK 73104 ([email protected]). 62 CUTIS® WWW.CUTIS.COM Copyright Cutis 2012. No part of this publication may be reproduced, stored, or transmitted without the prior written permission of the Publisher. Photo Quiz Discussion The Diagnosis: Demodex Folliculitis CUTIS he most common ectoparasites in humans an increased number of sebaceous glands, such as the are Demodex mites.1 The mite Demodex face, scalp, neck, eyelids, and upper chest.7 In most Tfolliculorum was first discovered in cerumen cases, the presence of these mites is asymptomatic in 1841 by the anatomist Jakob Henle; the mite was and causes no clinical findings. -

Mites of Public Health Importance And
MITES OF PUBLIC HEALTH IMPORTANCE AND THEIR CONTROL TRAINING GUIDE - INSECT CONTROL SERIES Harry D. Pratt U. S. D E P A R T M E N T OF H E A L T H , EDU CA TIO N , A N D W E L F A R E PUBLIC HEALTH SERVICE Communicable Disease Center Atlanta, Georgia Names of commercial mataiifacturers and trade names are provided as example only, and their inclusion does not imply endorsement by the Public Health Service or the U. S. Department of Health, Education, and Welfare; nor does the ex clusion of other commercial manufacturers and trade names imply nonendorsement by the Service or Department. The following titles in the Insect Control Series, Public Health Service Publication No.772, have been published. A ll are on sale at the Superintendent of Documents, Washington 25, D .C., at the prices shown: Part I: Introduction to Arthropods of Public Health Importance, 30 cents Part II: Insecticides for the Control of Insects of Public Health Importance, 30 cents Part III: Insecticidal Equipment for the Control of Insects of Public Health Importance, 25 cents Part IV: Sanitation in the Control of Insects and Rodents of Public Health Importance, 35 cents Part V: Flies of Public Health Importance and Their Control, 30 cents Part VII: Fleas of Public Health Importance and Their Control, 30 cents Part VIII: Lice of Public Health Importance and Their Control, 20 cents Part X: Ticks of Public Health Importance and Their Control, 30 cents These additional parts will appear at intervals: Part VI: Mosquitoes of Public Health Importance and Their Control Part XI: Scorpions, Spiders and Other Arthropods of Minor Public Health Importance and Their Control Part XII: Household and Stored-Food Insects of Public Health Importance and Their Control Public Health Service Publication No.772 Insect Control Series: Part IX May 1963 UNITED STATES GOVERNMENT PRINTING OFFICE, WASHINGTON: 1963 For sale by the Superintendent of Documents, U.S.