Details of Published Sequences of 18S Rdna Used in Phylogenetic Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Basal Body Structure and Composition in the Apicomplexans Toxoplasma and Plasmodium Maria E
Francia et al. Cilia (2016) 5:3 DOI 10.1186/s13630-016-0025-5 Cilia REVIEW Open Access Basal body structure and composition in the apicomplexans Toxoplasma and Plasmodium Maria E. Francia1* , Jean‑Francois Dubremetz2 and Naomi S. Morrissette3 Abstract The phylum Apicomplexa encompasses numerous important human and animal disease-causing parasites, includ‑ ing the Plasmodium species, and Toxoplasma gondii, causative agents of malaria and toxoplasmosis, respectively. Apicomplexans proliferate by asexual replication and can also undergo sexual recombination. Most life cycle stages of the parasite lack flagella; these structures only appear on male gametes. Although male gametes (microgametes) assemble a typical 9 2 axoneme, the structure of the templating basal body is poorly defined. Moreover, the rela‑ tionship between asexual+ stage centrioles and microgamete basal bodies remains unclear. While asexual stages of Plasmodium lack defined centriole structures, the asexual stages of Toxoplasma and closely related coccidian api‑ complexans contain centrioles that consist of nine singlet microtubules and a central tubule. There are relatively few ultra-structural images of Toxoplasma microgametes, which only develop in cat intestinal epithelium. Only a subset of these include sections through the basal body: to date, none have unambiguously captured organization of the basal body structure. Moreover, it is unclear whether this basal body is derived from pre-existing asexual stage centrioles or is synthesized de novo. Basal bodies in Plasmodium microgametes are thought to be synthesized de novo, and their assembly remains ill-defined. Apicomplexan genomes harbor genes encoding δ- and ε-tubulin homologs, potentially enabling these parasites to assemble a typical triplet basal body structure. -
Molecular Data and the Evolutionary History of Dinoflagellates by Juan Fernando Saldarriaga Echavarria Diplom, Ruprecht-Karls-Un
Molecular data and the evolutionary history of dinoflagellates by Juan Fernando Saldarriaga Echavarria Diplom, Ruprecht-Karls-Universitat Heidelberg, 1993 A THESIS SUBMITTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in THE FACULTY OF GRADUATE STUDIES Department of Botany We accept this thesis as conforming to the required standard THE UNIVERSITY OF BRITISH COLUMBIA November 2003 © Juan Fernando Saldarriaga Echavarria, 2003 ABSTRACT New sequences of ribosomal and protein genes were combined with available morphological and paleontological data to produce a phylogenetic framework for dinoflagellates. The evolutionary history of some of the major morphological features of the group was then investigated in the light of that framework. Phylogenetic trees of dinoflagellates based on the small subunit ribosomal RNA gene (SSU) are generally poorly resolved but include many well- supported clades, and while combined analyses of SSU and LSU (large subunit ribosomal RNA) improve the support for several nodes, they are still generally unsatisfactory. Protein-gene based trees lack the degree of species representation necessary for meaningful in-group phylogenetic analyses, but do provide important insights to the phylogenetic position of dinoflagellates as a whole and on the identity of their close relatives. Molecular data agree with paleontology in suggesting an early evolutionary radiation of the group, but whereas paleontological data include only taxa with fossilizable cysts, the new data examined here establish that this radiation event included all dinokaryotic lineages, including athecate forms. Plastids were lost and replaced many times in dinoflagellates, a situation entirely unique for this group. Histones could well have been lost earlier in the lineage than previously assumed. -

Parasitic Dinoflagellate Hematodinium Perezi Prevalence in Larval and Juvenile Blue Crabs Callinectes Sapidus from Coastal Bays of Virginia
W&M ScholarWorks VIMS Articles Virginia Institute of Marine Science 6-6-2019 Parasitic dinoflagellate Hematodinium perezi prevalence in larval and juvenile blue crabs Callinectes sapidus from coastal bays of Virginia HJ Small Virginia Institute of Marine Science JP Huchin-Mian Virginia Institute of Marine Science KS Reece Virginia Institute of Marine Science KM Pagenkopp Lohan MJ Butler IV See next page for additional authors Follow this and additional works at: https://scholarworks.wm.edu/vimsarticles Part of the Marine Biology Commons, and the Parasitology Commons Recommended Citation Small, HJ; Huchin-Mian, JP; Reece, KS; Pagenkopp Lohan, KM; Butler, MJ IV; and Shields, JD, Parasitic dinoflagellate Hematodinium perezi prevalence in larval and juvenile blue crabs Callinectes sapidus from coastal bays of Virginia (2019). Diseases of Aquatic Organisms, 134, 215-222. 10.3354/dao03371 This Article is brought to you for free and open access by the Virginia Institute of Marine Science at W&M ScholarWorks. It has been accepted for inclusion in VIMS Articles by an authorized administrator of W&M ScholarWorks. For more information, please contact [email protected]. Authors HJ Small, JP Huchin-Mian, KS Reece, KM Pagenkopp Lohan, MJ Butler IV, and JD Shields This article is available at W&M ScholarWorks: https://scholarworks.wm.edu/vimsarticles/1428 Vol. 134: 215–222, 2019 DISEASES OF AQUATIC ORGANISMS Published online June 6 https://doi.org/10.3354/dao03371 Dis Aquat Org OPENPEN ACCESSCCESS Parasitic dinoflagellate Hematodinium perezi prevalence in larval and juvenile blue crabs Callinectes sapidus from coastal bays of Virginia H. J. Small1,*, J. P. Huchin-Mian1,3, K. -

The Planktonic Protist Interactome: Where Do We Stand After a Century of Research?
bioRxiv preprint doi: https://doi.org/10.1101/587352; this version posted May 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Bjorbækmo et al., 23.03.2019 – preprint copy - BioRxiv The planktonic protist interactome: where do we stand after a century of research? Marit F. Markussen Bjorbækmo1*, Andreas Evenstad1* and Line Lieblein Røsæg1*, Anders K. Krabberød1**, and Ramiro Logares2,1** 1 University of Oslo, Department of Biosciences, Section for Genetics and Evolutionary Biology (Evogene), Blindernv. 31, N- 0316 Oslo, Norway 2 Institut de Ciències del Mar (CSIC), Passeig Marítim de la Barceloneta, 37-49, ES-08003, Barcelona, Catalonia, Spain * The three authors contributed equally ** Corresponding authors: Ramiro Logares: Institute of Marine Sciences (ICM-CSIC), Passeig Marítim de la Barceloneta 37-49, 08003, Barcelona, Catalonia, Spain. Phone: 34-93-2309500; Fax: 34-93-2309555. [email protected] Anders K. Krabberød: University of Oslo, Department of Biosciences, Section for Genetics and Evolutionary Biology (Evogene), Blindernv. 31, N-0316 Oslo, Norway. Phone +47 22845986, Fax: +47 22854726. [email protected] Abstract Microbial interactions are crucial for Earth ecosystem function, yet our knowledge about them is limited and has so far mainly existed as scattered records. Here, we have surveyed the literature involving planktonic protist interactions and gathered the information in a manually curated Protist Interaction DAtabase (PIDA). In total, we have registered ~2,500 ecological interactions from ~500 publications, spanning the last 150 years. -

University of Oklahoma
UNIVERSITY OF OKLAHOMA GRADUATE COLLEGE MACRONUTRIENTS SHAPE MICROBIAL COMMUNITIES, GENE EXPRESSION AND PROTEIN EVOLUTION A DISSERTATION SUBMITTED TO THE GRADUATE FACULTY in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY By JOSHUA THOMAS COOPER Norman, Oklahoma 2017 MACRONUTRIENTS SHAPE MICROBIAL COMMUNITIES, GENE EXPRESSION AND PROTEIN EVOLUTION A DISSERTATION APPROVED FOR THE DEPARTMENT OF MICROBIOLOGY AND PLANT BIOLOGY BY ______________________________ Dr. Boris Wawrik, Chair ______________________________ Dr. J. Phil Gibson ______________________________ Dr. Anne K. Dunn ______________________________ Dr. John Paul Masly ______________________________ Dr. K. David Hambright ii © Copyright by JOSHUA THOMAS COOPER 2017 All Rights Reserved. iii Acknowledgments I would like to thank my two advisors Dr. Boris Wawrik and Dr. J. Phil Gibson for helping me become a better scientist and better educator. I would also like to thank my committee members Dr. Anne K. Dunn, Dr. K. David Hambright, and Dr. J.P. Masly for providing valuable inputs that lead me to carefully consider my research questions. I would also like to thank Dr. J.P. Masly for the opportunity to coauthor a book chapter on the speciation of diatoms. It is still such a privilege that you believed in me and my crazy diatom ideas to form a concise chapter in addition to learn your style of writing has been a benefit to my professional development. I’m also thankful for my first undergraduate research mentor, Dr. Miriam Steinitz-Kannan, now retired from Northern Kentucky University, who was the first to show the amazing wonders of pond scum. Who knew that studying diatoms and algae as an undergraduate would lead me all the way to a Ph.D. -

ABSTRACT Gregarine Parasitism in Dragonfly Populations of Central
ABSTRACT Gregarine Parasitism in Dragonfly Populations of Central Texas with an Assessment of Fitness Costs in Erythemis simplicicollis Jason L. Locklin, Ph.D. Mentor: Darrell S. Vodopich, Ph.D. Dragonfly parasites are widespread and frequently include gregarines (Phylum Apicomplexa) in the gut of the host. Gregarines are ubiquitous protozoan parasites that infect arthropods worldwide. More than 1,600 gregarine species have been described, but only a small percentage of invertebrates have been surveyed for these apicomplexan parasites. Some consider gregarines rather harmless, but recent studies suggest otherwise. Odonate-gregarine studies have more commonly involved damselflies, and some have considered gregarines to rarely infect dragonflies. In this study, dragonfly populations were surveyed for gregarines and an assessment of fitness costs was made in a common and widespread host species, Erythemis simplicicollis. Adult dragonfly populations were surveyed weekly at two reservoirs in close proximity to one another and at a flow-through wetland system. Gregarine prevalences and intensities were compared within host populations between genders, among locations, among wing loads, and through time. Host fitness parameters measured included wing load, egg size, clutch size, and total egg count. Of the 37 dragonfly species surveyed, 14 species (38%) hosted gregarines. Thirteen of those species were previously unreported as hosts. Gregarine prevalences ranged from 2% – 52%. Intensities ranged from 1 – 201. Parasites were aggregated among their hosts. Gregarines were found only in individuals exceeding a minimum wing load, indicating that gregarines are likely not transferred from the naiad to adult during emergence. Prevalence and intensity exhibited strong seasonality during both years at one of the reservoirs, but no seasonal trend was detected at the wetland. -

Symbiodinium Genomes Reveal Adaptive Evolution of Functions Related to Symbiosis
bioRxiv preprint doi: https://doi.org/10.1101/198762; this version posted October 5, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Article 2 Symbiodinium genomes reveal adaptive evolution of 3 functions related to symbiosis 4 Huanle Liu1, Timothy G. Stephens1, Raúl A. González-Pech1, Victor H. Beltran2, Bruno 5 Lapeyre3,4, Pim Bongaerts5, Ira Cooke3, David G. Bourne2,6, Sylvain Forêt7,*, David J. 6 Miller3, Madeleine J. H. van Oppen2,8, Christian R. Voolstra9, Mark A. Ragan1 and Cheong 7 Xin Chan1,10,† 8 1Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD 4072, 9 Australia 10 2Australian Institute of Marine Science, Townsville, QLD 4810, Australia 11 3ARC Centre of Excellence for Coral Reef Studies and Department of Molecular and Cell 12 Biology, James Cook University, Townsville, QLD 4811, Australia 13 4Laboratoire d’excellence CORAIL, Centre de Recherches Insulaires et Observatoire de 14 l’Environnement, Moorea 98729, French Polynesia 15 5Global Change Institute, The University of Queensland, Brisbane, QLD 4072, Australia 16 6College of Science and Engineering, James Cook University, Townsville, QLD 4811, 17 Australia 18 7Research School of Biology, Australian National University, Canberra, ACT 2601, Australia 19 8School of BioSciences, The University of Melbourne, VIC 3010, Australia 1 bioRxiv preprint doi: https://doi.org/10.1101/198762; this version posted October 5, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Structure of Protistan Parasites Found in Bivalve Molluscs
W&M ScholarWorks VIMS Books and Book Chapters Virginia Institute of Marine Science 1988 Structure of Protistan Parasites Found in Bivalve Molluscs Frank O. Perkins Follow this and additional works at: https://scholarworks.wm.edu/vimsbooks Part of the Marine Biology Commons, and the Parasitology Commons American Fisheries Society Special Publication 18:93- 111 , 1988 CC> Copyrighl by !he American Fisheries Sociely 1988 PARASITE MORPHOLOGY, STRATEGY, AND EVOLUTION Structure of Protistan Parasites Found in Bivalve Molluscs 1 FRANK 0. PERKINS Virginia In stitute of Marine Science. School of Marine Science, College of William and Mary Gloucester Point, Virginia 23062, USA Abstral'I.-The literature on the structure of protists parasitizing bivalve molluscs is reviewed, and previously unpubli shed observations of species of class Perkinsea, phylum Haplosporidia, and class Paramyxea are presented. Descriptions are given of the flagellar apparatus of Perkin.His marinus zoospores, the ultrastructure of Perkinsus sp. from the Baltic macoma Maconw balthica, and the development of haplosporosome-like bodies in Haplosporidium nelsoni. The possible origin of stem cells of Marreilia sydneyi from the inner two sporoplasms is discussed. New research efforts are suggested which could help elucidate the phylogenetic interrelationships and taxonomic positions of the various taxa and help in efforts to better understand life cycles of selected species. Studies of the structure of protistan parasites terization of the parasite species, to elucidation of found in bivalve moll uscs have been fruitful to the many parasite life cycles, and to knowledge of morphologist interested in comparative morphol- parasite metabolism. The latter, especially, is ogy, evolu tion, and taxonomy. -

Metagenomic Characterization of Unicellular Eukaryotes in the Urban Thessaloniki Bay
Metagenomic characterization of unicellular eukaryotes in the urban Thessaloniki Bay George Tsipas SCHOOL OF ECONOMICS, BUSINESS ADMINISTRATION & LEGAL STUDIES A thesis submitted for the degree of Master of Science (MSc) in Bioeconomy Law, Regulation and Management May, 2019 Thessaloniki – Greece George Tsipas ’’Metagenomic characterization of unicellular eukaryotes in the urban Thessaloniki Bay’’ Student Name: George Tsipas SID: 268186037282 Supervisor: Prof. Dr. Savvas Genitsaris I hereby declare that the work submitted is mine and that where I have made use of another’s work, I have attributed the source(s) according to the Regulations set in the Student’s Handbook. May, 2019 Thessaloniki - Greece Page 2 of 63 George Tsipas ’’Metagenomic characterization of unicellular eukaryotes in the urban Thessaloniki Bay’’ 1. Abstract The present research investigates through metagenomics sequencing the unicellular protistan communities in Thermaikos Gulf. This research analyzes the diversity, composition and abundance in this marine environment. Water samples were collected monthly from April 2017 to February 2018 in the port of Thessaloniki (Harbor site, 40o 37’ 55 N, 22o 56’ 09 E). The extraction of DNA was completed as well as the sequencing was performed, before the downstream read processing and the taxonomic classification that was assigned using PR2 database. A total of 1248 Operational Taxonomic Units (OTUs) were detected but only 700 unicellular eukaryotes were analyzed, excluding unclassified OTUs, Metazoa and Streptophyta. In this research-based study the most abundant and diverse taxonomic groups were Dinoflagellata and Protalveolata. Specifically, the most abundant groups of all samples are Dinoflagellata with 190 OTUs (27.70%), Protalveolata with 139 OTUs (20.26%) Ochrophyta with 73 OTUs (10.64%), Cercozoa with 67 OTUs (9.77%) and Ciliophora with 64 OTUs (9.33%). -
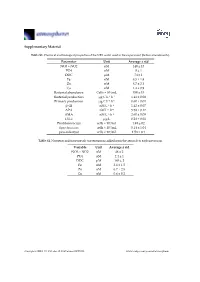
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

Resource Partitioning Between Phytoplankton and Bacteria in the Coastal Baltic Sea Frontiers in Marine Science, 7: 1-19
http://www.diva-portal.org This is the published version of a paper published in Frontiers in Marine Science. Citation for the original published paper (version of record): Sörenson, E., Lindehoff, E., Farnelid, H., Legrand, C. (2020) Resource Partitioning Between Phytoplankton and Bacteria in the Coastal Baltic Sea Frontiers in Marine Science, 7: 1-19 https://doi.org/10.3389/fmars.2020.608244 Access to the published version may require subscription. N.B. When citing this work, cite the original published paper. Permanent link to this version: http://urn.kb.se/resolve?urn=urn:nbn:se:lnu:diva-99520 ORIGINAL RESEARCH published: 25 November 2020 doi: 10.3389/fmars.2020.608244 Resource Partitioning Between Phytoplankton and Bacteria in the Coastal Baltic Sea Eva Sörenson, Hanna Farnelid, Elin Lindehoff and Catherine Legrand* Department of Biology and Environmental Science, Linnaeus University Centre of Ecology and Evolution and Microbial Model Systems, Linnaeus University, Kalmar, Sweden Eutrophication coupled to climate change disturbs the balance between competition and coexistence in microbial communities including the partitioning of organic and inorganic nutrients between phytoplankton and bacteria. Competition for inorganic nutrients has been regarded as one of the drivers affecting the productivity of the eutrophied coastal Baltic Sea. Yet, it is unknown at the molecular expression level how resources are competed for, by phytoplankton and bacteria, and what impact this competition has on the community composition. Here we use metatranscriptomics and amplicon sequencing and compare known metabolic pathways of both phytoplankton and bacteria co-occurring during a summer bloom in the archipelago of Åland in the Baltic Sea to examine phytoplankton bacteria resource partitioning. -

PROTISTAS MARINOS Viviana A
PROTISTAS MARINOS Viviana A. Alder INTRODUCCIÓN plantas y animales. Según este esquema básico, a las plantas les correspondían las características de En 1673, el editor de Philosophical Transac- ser organismos sésiles con pigmentos fotosinté- tions of the Royal Society of London recibió una ticos para la síntesis de las sustancias esenciales carta del anatomista Regnier de Graaf informan- para su metabolismo a partir de sustancias inor- do que un comerciante holandés, Antonie van gánicas (nutrición autótrofa), y de poseer células Leeuwenhoek, había “diseñado microscopios rodeadas por paredes de celulosa. En oposición muy superiores a aquéllos que hemos visto has- a las plantas, les correspondía a los animales los ta ahora”. Van Leeuwenhoek vendía lana, algo- atributos de tener motilidad activa y de carecer dón y otros materiales textiles, y se había visto tanto de pigmentos fotosintéticos (debiendo por en la necesidad de mejorar las lentes de aumento lo tanto procurarse su alimento a partir de sustan- que comúnmente usaba para contar el número cias orgánicas sintetizadas por otros organismos) de hebras y evaluar la calidad de fibras y tejidos. como de paredes celulósicas en sus células. Así fue que construyó su primer microscopio de Es a partir de los estudios de Georg Gol- lente única: simple, pequeño, pero con un poder dfuss (1782-1848) que estos diminutos organis- de magnificación de hasta 300 aumentos (¡diez mos, invisibles a ojo desnudo, comienzan a ser veces más que sus precursores!). Este magnífico clasificados como plantas primarias