Nucleotide Exchange Factors Kinetic Analyses and the Rationale for Studying Kinetics of Gefs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hras Intracellular Trafficking and Signal Transduction Jodi Ho-Jung Mckay Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2007 HRas intracellular trafficking and signal transduction Jodi Ho-Jung McKay Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cancer Biology Commons, Cell Biology Commons, Genetics and Genomics Commons, and the Medical Cell Biology Commons Recommended Citation McKay, Jodi Ho-Jung, "HRas intracellular trafficking and signal transduction" (2007). Retrospective Theses and Dissertations. 13946. https://lib.dr.iastate.edu/rtd/13946 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. HRas intracellular trafficking and signal transduction by Jodi Ho-Jung McKay A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Genetics Program of Study Committee: Janice E. Buss, Co-major Professor Linda Ambrosio, Co-major Professor Diane Bassham Drena Dobbs Ted Huiatt Iowa State University Ames, Iowa 2007 Copyright © Jodi Ho-Jung McKay, 2007. All rights reserved. UMI Number: 3274881 Copyright 2007 by McKay, Jodi Ho-Jung All rights reserved. UMI Microform 3274881 Copyright 2008 by ProQuest Information and Learning Company. All rights reserved. This microform edition is protected against unauthorized copying under Title 17, United States Code. ProQuest Information and Learning Company 300 North Zeeb Road P.O. -

ADP-Ribosylation Factor, a Small GTP-Binding Protein, Is Required for Binding of the Coatomer Protein Fl-COP to Golgi Membranes JULIE G
Proc. Natl. Acad. Sci. USA Vol. 89, pp. 6408-6412, July 1992 Biochemistry ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein fl-COP to Golgi membranes JULIE G. DONALDSON*, DAN CASSEL*t, RICHARD A. KAHN*, AND RICHARD D. KLAUSNER* *Cell Biology and Metabolism Branch, National Institute of Child Health and Human Development, and tLaboratory of Biological Chemistry, Division of Cancer Treatment, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892 Communicated by Marc Kirschner, April 20, 1992 (receivedfor review February 11, 1992) ABSTRACT The coatomer is a cytosolic protein complex localized to the Golgi complex, although their functions have that reversibly associates with Golgi membranes and is Impli- not been defined. Distinct among these proteins is the ADP- cated in modulating Golgi membrane transport. The associa- ribosylation factor (ARF), originally identified as a cofactor tion of 13-COP, a component of coatomer, with Golgi mem- required for in vitro cholera toxin-catalyzed ADP- branes is enhanced by guanosine 5'-[v-thioltriphosphate ribosylation of the a subunit of the trimeric GTP-binding (GTP[yS]), a nonhydrolyzable analogue of GTP, and by a protein G, (G,.) (19). ARF is an abundant cytosolic protein mixture of aluminum and fluoride ions (Al/F). Here we show that reversibly associates with Golgi membranes (20, 21). that the ADP-ribosylation factor (ARF) is required for the ARF has been shown to be present on Golgi coated vesicles binding of (-COP. Thus, 13-COP contained in a coatomer generated in the presence of GTP[yS], but it is not a com- fraction that has been resolved from ARF does not bind to Golgi ponent of the cytosolic coatomer (22). -

Supplementary Table 2
Supplementary Table 2. Differentially Expressed Genes following Sham treatment relative to Untreated Controls Fold Change Accession Name Symbol 3 h 12 h NM_013121 CD28 antigen Cd28 12.82 BG665360 FMS-like tyrosine kinase 1 Flt1 9.63 NM_012701 Adrenergic receptor, beta 1 Adrb1 8.24 0.46 U20796 Nuclear receptor subfamily 1, group D, member 2 Nr1d2 7.22 NM_017116 Calpain 2 Capn2 6.41 BE097282 Guanine nucleotide binding protein, alpha 12 Gna12 6.21 NM_053328 Basic helix-loop-helix domain containing, class B2 Bhlhb2 5.79 NM_053831 Guanylate cyclase 2f Gucy2f 5.71 AW251703 Tumor necrosis factor receptor superfamily, member 12a Tnfrsf12a 5.57 NM_021691 Twist homolog 2 (Drosophila) Twist2 5.42 NM_133550 Fc receptor, IgE, low affinity II, alpha polypeptide Fcer2a 4.93 NM_031120 Signal sequence receptor, gamma Ssr3 4.84 NM_053544 Secreted frizzled-related protein 4 Sfrp4 4.73 NM_053910 Pleckstrin homology, Sec7 and coiled/coil domains 1 Pscd1 4.69 BE113233 Suppressor of cytokine signaling 2 Socs2 4.68 NM_053949 Potassium voltage-gated channel, subfamily H (eag- Kcnh2 4.60 related), member 2 NM_017305 Glutamate cysteine ligase, modifier subunit Gclm 4.59 NM_017309 Protein phospatase 3, regulatory subunit B, alpha Ppp3r1 4.54 isoform,type 1 NM_012765 5-hydroxytryptamine (serotonin) receptor 2C Htr2c 4.46 NM_017218 V-erb-b2 erythroblastic leukemia viral oncogene homolog Erbb3 4.42 3 (avian) AW918369 Zinc finger protein 191 Zfp191 4.38 NM_031034 Guanine nucleotide binding protein, alpha 12 Gna12 4.38 NM_017020 Interleukin 6 receptor Il6r 4.37 AJ002942 -

ADP Ribosylation Factor 6 Is Activated and Controls Membrane Delivery
JCBArticle ADP ribosylation factor 6 is activated and controls membrane delivery during phagocytosis in macrophages Florence Niedergang,1 Emma Colucci-Guyon,1 Thierry Dubois,1 Graça Raposo,2 and Philippe Chavrier1 1Membrane and Cytoskeleton Dynamics Group and 2Electron Microscopy Group, UMR 144 Centre National de la Recherche Scientifique, Institut Curie, F-75248 Paris Cedex 05, France ngulfment of particles by phagocytes is induced by immunoglobulins (FcRs). A dominant-negative mutant of their interaction with specific receptors on the cell ARF6 (T27N mutation) dramatically affected FcR-mediated Esurface, which leads to actin polymerization and the phagocytosis. Expression of ARF6-T27N lead to a reduction extension of membrane protrusions to form a closed phago- in the focal delivery of vesicle-associated membrane protein some. Membrane delivery from internal pools is considered 3ϩ endosomal recycling membranes at phagocytosis sites, to play an important role in pseudopod extension during whereas actin polymerization was unimpaired. This resulted phagocytosis. Here, we report that endogenous ADP ribosyla- in an early blockade in pseudopod extension and accumu- tion factor 6 (ARF6), a small GTP-binding protein, undergoes lation of intracellular vesicles, as observed by electron a sharp and transient activation in macrophages when phago- microscopy. We conclude that ARF6 is a major regulator of cytosis was initiated via receptors for the Fc portion of membrane recycling during phagocytosis. Introduction Phagocytosis is the mechanism of internalization used by cells the plasma membrane to be locally elongated to form the to take up relatively large particles (Ͼ0.5 m) into an intra- engulfing pseudopods (Castellano et al., 2001; May and cellular compartment or phagosome. -

Studies on the Inhibition of Endosome Fusion by Gtpγs-Bound ARF
Journal of Cell Science 112, 3477-3485 (1999) 3477 Printed in Great Britain © The Company of Biologists Limited 1999 JCS0438 Studies on the inhibition of endosome fusion by GTPγS-bound ARF Arwyn T. Jones1, David J. Spiro1, Tomas Kirchhausen2, Paul Melançon3 and Marianne Wessling-Resnick1,* 1Department of Nutrition, Harvard School of Public Health, Boston, MA 02115, USA 2Department of Cell Biology, Harvard Medical School, Center for Blood Research, Boston, MA 02115, USA 3Department of Cell Biology, University of Alberta, Edmonton, AB T6G 2H7, Canada *Author for correspondence (e-mail: [email protected]) Accepted 28 July; published on WWW 30 September 1999 SUMMARY Using a cell free assay, we have previously shown that ARF independent of endosomal acidification since assays is not required for endosome fusion but that inhibition of performed in the presence of the vacuolar ATPase inhibitor fusion by GTPγS is dependent on a cytosolic pool of ARFs. bafilomycin A1 are equally sensitive to GTPγS-bound ARF. Since ARF is proposed to function in intracellular Finally, in contrast to reported effects on lysosomes, we membrane traffic by promoting vesicle biogenesis, and demonstrate that ARF-GTPγS does not induce endosomal components of clathrin- and COP-coated vesicles have been lysis. These combined data argue that sequestration of localized on endosomal structures, we investigated whether known coat proteins to membranes by activated ARF is not ARF-mediated inhibition of early endosome fusion involves involved in the inhibition of early endosome fusion and that the recruitment or irreversible association of these proteins its capacity to inhibit fusion involves other specific onto endosomal membranes. -

A Presynaptic Role for the ADP Ribosylation Factor (ARF)-Specific GDP͞GTP Exchange Factor Msec7-1
Proc. Natl. Acad. Sci. USA Vol. 96, pp. 1094–1099, February 1999 Neurobiology A presynaptic role for the ADP ribosylation factor (ARF)-specific GDPyGTP exchange factor msec7-1 URI ASHERY*†,HENRIETTE KOCH†‡,VOLKER SCHEUSS*, NILS BROSE‡, AND JENS RETTIG*§ *Department of Membrane Biophysics, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, D-37077 Goettingen, Germany; and ‡Department of Neurobiology, Max Planck Institute for Experimental Medicine, Hermann-Rein-Strasse 3, D-37075 Goettingen, Germany Communicated by Erwin Neher, Max Planck Institute for Biophysical Chemistry, Goettingen, Germany, November 30, 1998 (received for review September 15, 1998) ABSTRACT ADP ribosylation factors (ARFs) represent a cycle. In the axon terminal, transmitter is released by fusion of family of small monomeric G proteins that switch from an synaptic vesicles with the plasma membrane. Synaptic vesicles inactive, GDP-bound state to an active, GTP-bound state. One are formed by budding from early endosomal compartments. member of this family, ARF6, translocates on activation from They then are filled with neurotransmitter and translocate to intracellular compartments to the plasma membrane and has a specialized region of the plasma membrane, the active zone, been implicated in regulated exocytosis in neuroendocrine where they dock and mature to a fusion competent state. cells. Because GDP release in vivo is rather slow, ARF acti- Vesicles then fuse with the plasma membrane in response to vation is facilitated by specific guanine nucleotide exchange an elevated intracellular calcium concentration (e.g., after an factors like cytohesin-1 or ARNO. Here we show that msec7-1, action potential). Vesicular protein and lipid components are a rat homologue of cytohesin-1, translocates ARF6 to the retrieved by clathrin-mediated endocytosis and are recycled plasma membrane in living cells. -

Transcriptome-Wide Analysis of CXCR5 Deficient Retinal Pigment
Article Transcriptome‐wide analysis of CXCR5 deficient retinal pigment epithelial (RPE) cells reveals molecular signature of RPE homeostasis Supplementary data Madhu Sudhana Saddala 1, Anton Lennikov 1, Anthony Mukwaya 2, and Hu Huang 1,* 1 Department of Ophthalmology, University of Missouri, Columbia, MO 65212, United States of America; [email protected] (M.S.S.); [email protected] (A.L.) 2 Department of Ophthalmology, Institute for Clinical and Experimental Medicine, Faculty of Health Sciences, Linköping University, Linköping, 58183 , Sweden; [email protected] * Correspondence: [email protected]; Tel: +1 573‐882‐9899 # Madhu Sudhana Saddala and Anton Lennikov have contributed equally to this work. Keywords: Age‐related macular degeneration; CXCR5; EMT, FoxO; Mitochondria; RNA‐Seq, Gene Ontology; KEGG; Retinal pigment epithelium Supplementary Figures Biomedicines 2020, 8, x; doi: FOR PEER REVIEW www.mdpi.com/journal/biomedicines Biomolecules 2020, 10, x FOR PEER REVIEW 2 of 13 Biomolecules 2020, 10, x FOR PEER REVIEW 3 of 13 Figure S1: Quantitative profiling of mouse RPE tissue between control and CXCR5 ko groups. Pearson correlation coefficient plot of the log2 ratio between two groups (A) before and (B) after normalization. Potential relationships or correlations amongst the different data attributes is to leverage a pair‐wise correlation matrix in control and CXCR5 knockout groups. Biomolecules 2020, 10, x FOR PEER REVIEW 4 of 13 Biomolecules 2020, 10, x FOR PEER REVIEW 5 of 13 Figure S2: Potential relationships or correlations amongst the different data attributes is to leverage a pair‐wise correlation matrix in control and CXCR5 knockout groups. (A) heatmap of total differentially expressed genes (B) The cluster of main heatmap represented twenty differentially expressed genes. -
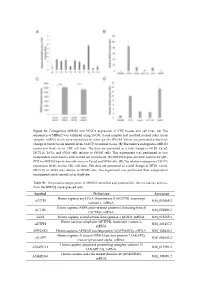
Figure S1. Endogenous MIR45
Figure S1. Endogenous MIR452 and VEGFA expression in CRC tissues and cell lines. (A) The expression of MIR452 was validated using 10 CRC tissue samples and matched normal colon tissue samples. miRNA levels were normalized to colon-specific RNU48. Values are presented as the fold- change in tumor tissue relative levels (ΔΔCT) to normal tissue. (B) The relative endogenous MIR452 expression levels in six CRC cell lines. The data are presented as a fold change in HT29, Caco2, HCT116, LoVo, and SW48 cells relative to SW480 cells. This experiment was performed as two independent experiments, each carried out in triplicate. (C) MIR452 expression level analysis by qRT- PCR for MIR452 transfection efficiency in Caco2 and SW48 cells. (D) The relative endogenous VEGFA expression levels in five CRC cell lines. The data are presented as a fold change in HT29, Caco2, HCT116, or SW48 cells relative to SW480 cells. This experiment was performed three independent experiments, each carried out in duplicate. Table S1. The putative target genes of MIR452 identified and predicted by the microarray analysis from the MIR452 overexpressed cells. Symbol Definition Accession Homo sapiens acyl-CoA thioesterase 8 (ACOT8), transcript ACOT8 NM_005469.2 variant 1, mRNA. Homo sapiens ARP6 actin-related protein 6 homolog (yeast) ACTR6 NM_022496.3 (ACTR6), mRNA. ADI1 Homo sapiens acireductone dioxygenase 1 (ADI1), mRNA. NM_018269.1 Homo sapiens aftiphilin (AFTPH), transcript variant 1, AFTPH NM_203437.2 mRNA. AHNAK2 Homo sapiens AHNAK nucleoprotein 2 (AHNAK2), mRNA. NM_138420.2 Homo sapiens A kinase (PRKA) anchor protein 7 (AKAP7), AKAP7 NM_004842.2 transcript variant alpha, mRNA. Homo sapiens anaphase promoting complex subunit 13 ANAPC13 NM_015391.2 (ANAPC13), mRNA. -

Role of Phospholipase D in G-Protein Coupled Receptor Function
Membranes 2014, 4, 302-318; doi:10.3390/membranes4030302 OPEN ACCESS membranes ISSN 2077-0375 www.mdpi.com/journal/membranes Review Role of Phospholipase D in G-Protein Coupled Receptor Function Lars-Ove Brandenburg 1,*, Thomas Pufe 1 and Thomas Koch 2 1 Department of Anatomy and Cell Biology, RWTH Aachen University, Wendlingweg 2, D-52074 Aachen, Germany; E-Mail: [email protected] 2 Department of Pharmacology and Toxicology, Otto-von-Guericke-University Magdeburg, D-39120 Magdeburg, Germany; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +49-241-808-9548; Fax: +49-241-808-2431. Received: 29 May 2014; in revised form: 24 June 2014 / Accepted: 25 June 2014 / Published: 3 July 2014 Abstract: Prolonged agonist exposure of many G-protein coupled receptors induces a rapid receptor phosphorylation and uncoupling from G-proteins. Resensitization of these desensitized receptors requires endocytosis and subsequent dephosphorylation. Numerous studies show the involvement of phospholipid-specific phosphodiesterase phospholipase D (PLD) in the receptor endocytosis and recycling of many G-protein coupled receptors e.g., opioid, formyl or dopamine receptors. The PLD hydrolyzes the headgroup of a phospholipid, generally phosphatidylcholine (PC), to phosphatidic acid (PA) and choline and is assumed to play an important function in cell regulation and receptor trafficking. Protein kinases and GTP binding proteins of the ADP-ribosylation and Rho families regulate the two mammalian PLD isoforms 1 and 2. Mammalian and yeast PLD are also potently stimulated by phosphatidylinositol 4,5-bisphosphate. The PA product is an intracellular lipid messenger. PLD and PA activities are implicated in a wide range of physiological processes and diseases including inflammation, diabetes, oncogenesis or neurodegeneration. -

ADP-Ribosylation Factor-Like 2 (ARL2) Regulates Cilia Stability and Development of Outer Segments in Rod Photoreceptor Neurons
Faculty Scholarship 2018 ADP-Ribosylation Factor-Like 2 (ARL2) regulates cilia stability and development of outer segments in rod photoreceptor neurons Zachary C. Wright West Virginia University Yuriy Loskutov West Virginia University Daniel Murphy West Virginia University Peter Stoilov West Virginia University Elena Pugacheva West Virginia University See next page for additional authors Follow this and additional works at: https://researchrepository.wvu.edu/faculty_publications Part of the Biochemistry Commons, Neurology Commons, and the Ophthalmology Commons Digital Commons Citation Wright, Zachary C.; Loskutov, Yuriy; Murphy, Daniel; Stoilov, Peter; Pugacheva, Elena; Goldberg, Andrew F.X.; and Ramamurthy, Visvanathan, "ADP-Ribosylation Factor-Like 2 (ARL2) regulates cilia stability and development of outer segments in rod photoreceptor neurons" (2018). Faculty Scholarship. 1273. https://researchrepository.wvu.edu/faculty_publications/1273 This Article is brought to you for free and open access by The Research Repository @ WVU. It has been accepted for inclusion in Faculty Scholarship by an authorized administrator of The Research Repository @ WVU. For more information, please contact [email protected]. Authors Zachary C. Wright, Yuriy Loskutov, Daniel Murphy, Peter Stoilov, Elena Pugacheva, Andrew F.X. Goldberg, and Visvanathan Ramamurthy This article is available at The Research Repository @ WVU: https://researchrepository.wvu.edu/faculty_publications/ 1273 www.nature.com/scientificreports OPEN ADP-Ribosylation Factor-Like 2 (ARL2) regulates cilia stability and development of outer segments in Received: 16 August 2018 Accepted: 28 October 2018 rod photoreceptor neurons Published: xx xx xxxx Zachary C. Wright1, Yuriy Loskutov2, Daniel Murphy2, Peter Stoilov2, Elena Pugacheva2, Andrew F. X. Goldberg 4 & Visvanathan Ramamurthy1,2,3 Photoreceptor cells are specialized neurons with a sensory cilium carrying an elaborate membrane structure, the outer segment (OS). -
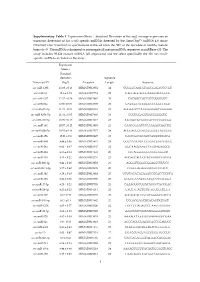
Supplementary Table 1. Expression
Supplementary Table 1. Expression (Mean Standard Deviation of the log2 average expression or transcript detection) of Sus scrofa specific miRNAs detected by the GeneChip™ miRNA 4.0 Array (ThermoFisher Scientific) in spermatozoa retrieved from the SRF of the ejaculate of healthy mature boars (n=3). The miRNA is designed to interrogate all mature miRNA sequences in miRBase v20. The array includes 30.424 mature miRNA (all organisms) and we select specifically the 326 Sus scrofa- specific miRNAs included in the array. Expression Mean ± Standard Deviation Sequence Transcript ID (log2) Accession Length Sequence ssc-miR-1285 13.98 ± 0.13 MIMAT0013954 24 CUGGGCAACAUAGCGAGACCCCGU ssc-miR-16 12.6 ± 0.74 MIMAT0007754 22 UAGCAGCACGUAAAUAUUGGCG ssc-miR-4332 12.32 ± 0.29 MIMAT0017962 20 CACGGCCGCCGCCGGGCGCC ssc-miR-92a 12.06 ± 0.09 MIMAT0013908 22 UAUUGCACUUGUCCCGGCCUGU ssc-miR-671-5p 11.73 ± 0.54 MIMAT0025381 24 AGGAAGCCCUGGAGGGGCUGGAGG ssc-miR-4334-5p 11.31 ± 0.05 MIMAT0017966 19 CCCUGGAGUGACGGGGGUG ssc-miR-425-5p 10.99 ± 0.15 MIMAT0013917 23 AAUGACACGAUCACUCCCGUUGA ssc-miR-191 10.57 ± 0.22 MIMAT0013876 23 CAACGGAAUCCCAAAAGCAGCUG ssc-miR-92b-5p 10.53 ± 0.18 MIMAT0017377 24 AGGGACGGGACGCGGUGCAGUGUU ssc-miR-15b 10.01 ± 0.9 MIMAT0002125 22 UAGCAGCACAUCAUGGUUUACA ssc-miR-30d 9.89 ± 0.36 MIMAT0013871 24 UGUAAACAUCCCCGACUGGAAGCU ssc-miR-26a 9.62 ± 0.47 MIMAT0002135 22 UUCAAGUAAUCCAGGAUAGGCU ssc-miR-484 9.55 ± 0.14 MIMAT0017974 20 CCCAGGGGGCGACCCAGGCU ssc-miR-103 9.53 ± 0.22 MIMAT0002154 23 AGCAGCAUUGUACAGGGCUAUGA ssc-miR-296-3p 9.41 ± 0.26 MIMAT0022958 -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN.