Role of Phospholipase D in G-Protein Coupled Receptor Function
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ATP-Induced Focal Adhesion Kinase Activity Is Negatively Modulated by Phospholipase D2 in PC12 Cells
EXPERIMENTAL and MOLECULAR MEDICINE, Vol. 33, No. 3, 150-155, September 2001 ATP-induced focal adhesion kinase activity is negatively modulated by phospholipase D2 in PC12 cells Yoe-Sik Bae1 and Sung Ho Ryu1,2 Introduction 1 Division of Molecular and Life Sciences, Pohang University of Purinergic receptors have been reported to play impor- Science and Technology, Pohang 790-784, Korea tant roles on the regulation of neuronal cell functions 2 Corresponding author: Tel, +82-54-279-2292; (Communi et al., 2000; Di Iorio et al., 1998). ATP, a Fax, +82-54-279-2199; E-mail, [email protected] ligand for the receptors modulate various cellular re- sponses such as mitogenic and morphogenic activity in Accepted 18 September 2001 PC12 rat pheochromocytoma cells (Neary et al., 1996; Soltoff et al., 1998; Schindelholz et al., 2000). Stimu- Abbreviations: Fak, focal adhesion kinase; PLD, phospholipase D; lation of cells with ATP induces tyrosine phosphorylation PA, phosphatidic acid; PC, phosphatidylcholine; DAG, diacylglyc- of several cytoskeletal proteins and focal adhesion erol; PBt, phosphatidylbutanol; PKC, protein kinase C; PAP, phos- molecules such as focal adhesion kinase (Fak), proline- phatidic acid phosphohydrolase rich tyrosine kinase (Pyk2), and paxillin (Soltoff et al., 1998; Schindelholz et al., 2000). Since these cytosk- eleton-associated proteins have been regarded as important factors for the regulation of neuronal cell Abstract functions, the study on the regulatory mechanism for the proteins remains an important issue. Extracellular ATP has been known to modulate vari- Phospholipase D (PLD) catalyzes the hydrolysis of ous cellular responses including mitogenesis, secre- phosphatidylcholine (PC) into phosphatidic acid (PA) tion and morphogenic activity in neuronal cells. -

Datasheet for Protein Phosphatase 1 (PP1) (P0754; Lot 0121306)
Supplied in: 200 mM NaCl, 50 mM HEPES Unit Definition: One unit is defined as Notes On Use: Avoid freeze/thaw cycles. Can be Protein the amount of enzyme that hydrolyzes 1 nmol of stored for 1 week or less at –20°C. (pH 7.0 @ 25°C), 1 mM MnCl2, 0.1 mM EGTA, Phosphatase 1 2.5 mM dithiothreitol, 0.025% Tween-20 and p-Nitrophenyl Phosphate (50 mM) (NEB #P0757) 50% glycerol. Store at –70°C in 1 minute at 30°C in a total reaction volume of The following information can be used as (PP1) 50 µl. suggested initial conditions for dephosphorylation 1-800-632-7799 Applications: PP1 can be used to release of proteins with PP1. [email protected] phosphate groups from phosphorylated serine, Specific Activity: ~ 80,000 units/mg. www.neb.com 0.1 unit of PP1 removes ~100% of phosphates P0754S 012130614061 threonine and tyrosine residues in proteins. Note that different proteins are dephosphorylated at Molecular Weight: 37.5 kDa. (0.5 nmol) from phosphoserine/threonine different rates. residues in phosphorylase a as well as in P0754S r y Purity: PP1 has been purified to > 90% phosphorylated myelin basic protein (phospho- 100 units 2,500 U/ml Lot: 0121306 Reagents Supplied with Enzyme: homogeneity as determined by SDS-PAGE and MyBP, 18.5 kDa) in 30 minutes in a 50 µl reaction. RECOMBINANT Store at –70°C Exp: 6/14 10X NEBuffer for Protein MetalloPhosphatases Coomassie Blue staining. The concentration of phospho-MyBP is 10 µM (PMP) with respect to phosphate. Description: Protein Phosphatase 1 (PP1) is a 10X MnCl2 (10 mM) Quality Assurance: PP1 contains no detectable Mn2+-dependent protein phosphatase with activity protease activity. -

Increased O-Glcnacylation Rapidly Decreases GABAAR Currents in Hippocampus but Depresses Neuronal Output L
www.nature.com/scientificreports OPEN Increased O-GlcNAcylation rapidly decreases GABAAR currents in hippocampus but depresses neuronal output L. T. Stewart1,3, K. Abiraman1,3, J. C. Chatham2 & L. L. McMahon1 ✉ O-GlcNAcylation, a post-translational modifcation involving O-linkage of β-N-acetylglucosamine to Ser/Thr residues on target proteins, is increasingly recognized as a critical regulator of synaptic function. Enzymes that catalyze O-GlcNAcylation are found at both presynaptic and postsynaptic sites, and O-GlcNAcylated proteins localize to synaptosomes. An acute increase in O-GlcNAcylation can afect neuronal communication by inducing long-term depression (LTD) of excitatory transmission at hippocampal CA3-CA1 synapses, as well as suppressing hyperexcitable circuits in vitro and in vivo. Despite these fndings, to date, no studies have directly examined how O-GlcNAcylation modulates the efcacy of inhibitory neurotransmission. Here we show an acute increase in O-GlcNAc dampens GABAergic currents onto principal cells in rodent hippocampus likely through a postsynaptic mechanism, and has a variable efect on the excitation/inhibition balance. The overall efect of increased O-GlcNAc is reduced synaptically-driven spike probability via synaptic depression and decreased intrinsic excitability. Our results position O-GlcNAcylation as a novel regulator of the overall excitation/inhibition balance and neuronal output. Synaptic integration and spike initiation in neurons is controlled by synaptic inhibition, which strongly infu- ences neuronal output and information processing1. Importantly, the balance of excitation to inhibition (E/I) is crucial to the proper functioning of circuits, and E/I imbalances have been implicated in a number of neurode- velopmental disorders and neurodegenerative diseases including schizophrenia, autism spectrum disorders, and Alzheimer’s disease2–5. -

Activation and Phosphorylation of the 'Dense-Vesicle' High-Affinity Cyclic AMP Phosphodiesterase by Cyclic AMP-Dependent Protein Kinase
Biochem. J. (1989) 260, 27-36 (Printed in Great Britain) 27 Activation and phosphorylation of the 'dense-vesicle' high-affinity cyclic AMP phosphodiesterase by cyclic AMP-dependent protein kinase Elaine KILGOUR, Neil G. ANDERSON and Miles D. HOUSLAY Molecular Pharmacology Group, Department of Biochemistry, University of Glasgow, Glasgow G12 8QQ, Scotland, U.K. Incubation of a hepatocyte particulate fraction with ATP and the isolated catalytic unit of cyclic AMP- dependent protein kinase (A-kinase) selectively activated the high-affinity 'dense-vesicle' cyclic AMP phosphodiesterase. Such activation only occurred if the membranes had been pre-treated with Mg2". Mg2" pre-treatment appeared to function by stimulating endogenous phosphatases and did not affect phosphodiesterase activity. Using the antiserum DV4, which specifically immunoprecipitated the 51 and 57 kDa components of the 'dense-vesicle' phosphodiesterase from a detergent-solubilized membrane extract, we isolated a 32P-labelled phosphoprotein from 32P-labelled hepatocytes. MgCl2 treatment of such labelled membranes removed 32P from the immunoprecipitated protein. Incubation of the Mg2+-pre-treated membranes with [32P]ATP and A-kinase led to the time-dependent incorporation of label into the 'dense- vesicle' phosphodiesterase, as detected by specific immunoprecipitation with the antiserum DV4. The time- dependences of phosphodiesterase activation and incorporation of label were similar. It is suggested (i) that phosphorylation of the 'dense-vesicle' phosphodiesterase by A-kinase leads to its activation, and that such a process accounts for the ability of glucagon and other hormones, which increase intracellular cyclic AMP concentrations, to activate this enzyme, and (ii) that an as yet unidentified kinase can phosphorylate this enzyme without causing any significant change in enzyme activity but which prevents activation and phosphorylation of the phosphodiesterase by A-kinase. -

Identification and Dynamics of the Human ZDHHC16-ZDHHC6 Palmitoylation Cascade
RESEARCH ARTICLE Identification and dynamics of the human ZDHHC16-ZDHHC6 palmitoylation cascade Laurence Abrami1†, Tiziano Dallavilla1,2†, Patrick A Sandoz1, Mustafa Demir1, Be´ atrice Kunz1, Georgios Savoglidis2, Vassily Hatzimanikatis2*, F Gisou van der Goot1* 1Global Health Institute, Faculty of Life Sciences, Ecole Polytechnique Fe´de´rale de Lausanne, Lausanne, Switzerland; 2Laboratory of Computational Systems Biotechnology, Faculty of Basic Sciences, Ecole Polytechnique Fe´de´rale de Lausanne, Lausanne, Switzerland Abstract S-Palmitoylation is the only reversible post-translational lipid modification. Knowledge about the DHHC palmitoyltransferase family is still limited. Here we show that human ZDHHC6, which modifies key proteins of the endoplasmic reticulum, is controlled by an upstream palmitoyltransferase, ZDHHC16, revealing the first palmitoylation cascade. The combination of site specific mutagenesis of the three ZDHHC6 palmitoylation sites, experimental determination of kinetic parameters and data-driven mathematical modelling allowed us to obtain detailed information on the eight differentially palmitoylated ZDHHC6 species. We found that species rapidly interconvert through the action of ZDHHC16 and the Acyl Protein Thioesterase APT2, that each species varies in terms of turnover rate and activity, altogether allowing the cell to robustly *For correspondence: tune its ZDHHC6 activity. [email protected] DOI: https://doi.org/10.7554/eLife.27826.001 (VH); [email protected] (FGG) †These authors contributed equally to this work Introduction Cells constantly interact with and respond to their environment. This requires tight control of protein Competing interests: The function in time and in space, which largely occurs through reversible post-translational modifica- authors declare that no tions of proteins, such as phosphorylation, ubiquitination and S-palmitoylation. -

Role of Phospholipases in Adrenal Steroidogenesis
229 1 W B BOLLAG Phospholipases in adrenal 229:1 R29–R41 Review steroidogenesis Role of phospholipases in adrenal steroidogenesis Wendy B Bollag Correspondence should be addressed Charlie Norwood VA Medical Center, One Freedom Way, Augusta, GA, USA to W B Bollag Department of Physiology, Medical College of Georgia, Augusta University (formerly Georgia Regents Email University), Augusta, GA, USA [email protected] Abstract Phospholipases are lipid-metabolizing enzymes that hydrolyze phospholipids. In some Key Words cases, their activity results in remodeling of lipids and/or allows the synthesis of other f adrenal cortex lipids. In other cases, however, and of interest to the topic of adrenal steroidogenesis, f angiotensin phospholipases produce second messengers that modify the function of a cell. In this f intracellular signaling review, the enzymatic reactions, products, and effectors of three phospholipases, f phospholipids phospholipase C, phospholipase D, and phospholipase A2, are discussed. Although f signal transduction much data have been obtained concerning the role of phospholipases C and D in regulating adrenal steroid hormone production, there are still many gaps in our knowledge. Furthermore, little is known about the involvement of phospholipase A2, Endocrinology perhaps, in part, because this enzyme comprises a large family of related enzymes of that are differentially regulated and with different functions. This review presents the evidence supporting the role of each of these phospholipases in steroidogenesis in the Journal Journal of Endocrinology adrenal cortex. (2016) 229, R1–R13 Introduction associated GTP-binding protein exchanges a bound GDP for a GTP. The G protein with GTP bound can then Phospholipids serve a structural function in the cell in that activate the enzyme, phospholipase C (PLC), that cleaves they form the lipid bilayer that maintains cell integrity. -

(PDE 1 B 1) Correlates with Brain Regions Having Extensive Dopaminergic Innervation
The Journal of Neuroscience, March 1994, 14(3): 1251-l 261 Expression of a Calmodulin-dependent Phosphodiesterase lsoform (PDE 1 B 1) Correlates with Brain Regions Having Extensive Dopaminergic Innervation Joseph W. Polli and Randall L. Kincaid Section on Immunology, Laboratory of Molecular and Cellular Neurobiology, National Institute on Alcohol Abuse and Alcoholism, Rockville, Maryland 20852 Cyclic nucleotide-dependent protein phosphorylation plays PDE implies an important physiological role for Ca2+-regu- a central role in neuronal signal transduction. Neurotrans- lated attenuation of CAMP-dependent signaling pathways mitter-elicited increases in cAMP/cGMP brought about by following dopaminergic stimulation. activation of adenylyl and guanylyl cyclases are downre- [Key words: CAMP, cyclase, striatum, dopamine, basal gulated by multiple phosphodiesterase (PDE) enzymes. In ganglia, DARPP-321 brain, the calmodulin (CaM)-dependent isozymes are the major degradative activities and represent a unique point of Cyclic nucleotides, acting as “second messengers”or via direct intersection between the cyclic nucleotide- and calcium effects, regulate a diverse array of neuronal functions, from ion (Ca*+)-mediated second messenger systems. Here we de- channel conductance to gene expression. Hydrolysis of 3’,5’- scribe the distribution of the PDEl Bl (63 kDa) CaM-depen- cyclic nucleotidesto 5’-nucleosidemonophosphates is the major dent PDE in mouse brain. An anti-peptide antiserum to this mechanismfor decreasingintracellular cyclic nucleotide levels. isoform immunoprecipitated -3O-40% of cytosolic PDE ac- This reaction is catalyzed by cyclic nucleotide phosphodiester- tivity, whereas antiserum to PDElA2 (61 kDa isoform) re- ase (PDE) enzymes that constitute a large superfamily (Beavo moved 60-70%, demonstrating that these isoforms are the and Reifsynder, 1990). -

Hras Intracellular Trafficking and Signal Transduction Jodi Ho-Jung Mckay Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2007 HRas intracellular trafficking and signal transduction Jodi Ho-Jung McKay Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cancer Biology Commons, Cell Biology Commons, Genetics and Genomics Commons, and the Medical Cell Biology Commons Recommended Citation McKay, Jodi Ho-Jung, "HRas intracellular trafficking and signal transduction" (2007). Retrospective Theses and Dissertations. 13946. https://lib.dr.iastate.edu/rtd/13946 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. HRas intracellular trafficking and signal transduction by Jodi Ho-Jung McKay A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Genetics Program of Study Committee: Janice E. Buss, Co-major Professor Linda Ambrosio, Co-major Professor Diane Bassham Drena Dobbs Ted Huiatt Iowa State University Ames, Iowa 2007 Copyright © Jodi Ho-Jung McKay, 2007. All rights reserved. UMI Number: 3274881 Copyright 2007 by McKay, Jodi Ho-Jung All rights reserved. UMI Microform 3274881 Copyright 2008 by ProQuest Information and Learning Company. All rights reserved. This microform edition is protected against unauthorized copying under Title 17, United States Code. ProQuest Information and Learning Company 300 North Zeeb Road P.O. -

A Novel, Highly Potent and Selective Phosphodiesterase-9 Inhibitor for the Ferrata Storti Foundation Treatment of Sickle Cell Disease
Red Cell Biology & its Disorders ARTICLE A novel, highly potent and selective phosphodiesterase-9 inhibitor for the Ferrata Storti Foundation treatment of sickle cell disease James G. McArthur,1 Niels Svenstrup,2 Chunsheng Chen,3 Aurelie Fricot,4 Caroline Carvalho,4 Julia Nguyen,3 Phong Nguyen,3 Anna Parachikova,2 Fuad Abdulla,3 Gregory M. Vercellotti,3 Olivier Hermine,4 Dave Edwards,5 Jean-Antoine Ribeil,6 John D. Belcher3 and Thiago T. Maciel4 1Imara Inc., 2nd Floor, 700 Technology Square, Cambridge, MA, USA; 2H. Lundbeck A/S, 3 Haematologica 2020 Ottiliavej 9, 2500 Valby, Denmark; Department of Medicine, Division of Hematology, Oncology and Transplantation, University of Minnesota, Minneapolis, MN, USA; Volume 105(3):623-631 4INSERM UMR 1163, CNRS ERL 8254, Imagine Institute, Laboratory of Excellence GR-Ex, Paris Descartes - Sorbonne Paris Cité University, Paris, France; 5Kinexum, 8830 Glen Ferry Drive, Johns Creek, GA, USA and 6Departments of Biotherapy, Necker Children’s Hospital, Assistance Publique-Hôpitaux de Paris (AP-HP), Paris Descartes- Sorbonne Paris Cité University, Paris, France ABSTRACT he most common treatment for patients with sickle cell disease (SCD) is the chemotherapeutic hydroxyurea, a therapy with Tpleiotropic effects, including increasing fetal hemoglobin (HbF) in red blood cells and reducing adhesion of white blood cells to the vascular endothelium. Hydroxyurea has been proposed to mediate these effects through a mechanism of increasing cellular cGMP levels. An alternative path to increasing cGMP levels in these cells is through the use of phospho- diesterase-9 inhibitors that selectively inhibit cGMP hydrolysis and increase cellular cGMP levels. We have developed a novel, potent and selective phosphodiesterase-9 inhibitor (IMR-687) specifically for the treatment of Correspondence: SCD. -

Regulation of Calmodulin-Stimulated Cyclic Nucleotide Phosphodiesterase (PDE1): Review
95-105 5/6/06 13:44 Page 95 INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 18: 95-105, 2006 95 Regulation of calmodulin-stimulated cyclic nucleotide phosphodiesterase (PDE1): Review RAJENDRA K. SHARMA, SHANKAR B. DAS, ASHAKUMARY LAKSHMIKUTTYAMMA, PONNIAH SELVAKUMAR and ANURAAG SHRIVASTAV Department of Pathology and Laboratory Medicine, College of Medicine, University of Saskatchewan, Cancer Research Division, Saskatchewan Cancer Agency, 20 Campus Drive, Saskatoon SK S7N 4H4, Canada Received January 16, 2006; Accepted March 13, 2006 Abstract. The response of living cells to change in cell 6. Differential inhibition of PDE1 isozymes and its environment depends on the action of second messenger therapeutic applications molecules. The two second messenger molecules cAMP and 7. Role of proteolysis in regulating PDE1A2 Ca2+ regulate a large number of eukaryotic cellular events. 8. Role of PDE1A1 in ischemic-reperfused heart Calmodulin-stimulated cyclic nucleotide phosphodiesterase 9. Conclusion (PDE1) is one of the key enzymes involved in the complex interaction between cAMP and Ca2+ second messenger systems. Some PDE1 isozymes have similar kinetic and 1. Introduction immunological properties but are differentially regulated by Ca2+ and calmodulin. Accumulating evidence suggests that the A variety of cellular activities are regulated through mech- activity of PDE1 is selectively regulated by cross-talk between anisms controlling the level of cyclic nucleotides. These Ca2+ and cAMP signalling pathways. These isozymes are mechanisms include synthesis, degradation, efflux and seque- also further distinguished by various pharmacological agents. stration of cyclic adenosine 3':5'-monophosphate (cAMP) and We have demonstrated a potentially novel regulation of PDE1 cyclic guanosine 3':5'- monophosphate (cGMP) within the by calpain. -

ADP-Ribosylation Factor, a Small GTP-Binding Protein, Is Required for Binding of the Coatomer Protein Fl-COP to Golgi Membranes JULIE G
Proc. Natl. Acad. Sci. USA Vol. 89, pp. 6408-6412, July 1992 Biochemistry ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein fl-COP to Golgi membranes JULIE G. DONALDSON*, DAN CASSEL*t, RICHARD A. KAHN*, AND RICHARD D. KLAUSNER* *Cell Biology and Metabolism Branch, National Institute of Child Health and Human Development, and tLaboratory of Biological Chemistry, Division of Cancer Treatment, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892 Communicated by Marc Kirschner, April 20, 1992 (receivedfor review February 11, 1992) ABSTRACT The coatomer is a cytosolic protein complex localized to the Golgi complex, although their functions have that reversibly associates with Golgi membranes and is Impli- not been defined. Distinct among these proteins is the ADP- cated in modulating Golgi membrane transport. The associa- ribosylation factor (ARF), originally identified as a cofactor tion of 13-COP, a component of coatomer, with Golgi mem- required for in vitro cholera toxin-catalyzed ADP- branes is enhanced by guanosine 5'-[v-thioltriphosphate ribosylation of the a subunit of the trimeric GTP-binding (GTP[yS]), a nonhydrolyzable analogue of GTP, and by a protein G, (G,.) (19). ARF is an abundant cytosolic protein mixture of aluminum and fluoride ions (Al/F). Here we show that reversibly associates with Golgi membranes (20, 21). that the ADP-ribosylation factor (ARF) is required for the ARF has been shown to be present on Golgi coated vesicles binding of (-COP. Thus, 13-COP contained in a coatomer generated in the presence of GTP[yS], but it is not a com- fraction that has been resolved from ARF does not bind to Golgi ponent of the cytosolic coatomer (22). -
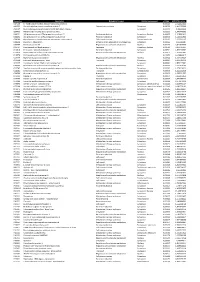
Accession Description Biological Process Cellular Component P-Value SCZ/CTRL Ration A4FU69 EF-Hand Calcium-Binding Domain-Contai
Accession Description Biological Process Cellular component p-Value SCZ/CTRL ration A4FU69 EF-hand calcium-binding domain-containing protein 5 0,001101 2,724427411 A4UGR9 Xin actin-binding repeat-containing protein 2 Cytoskeletal anchoring Cytoplasm 0,006756 1,413953388 A6NCE7 Microtubule-associated proteins 1A/1B light chain 3 beta 2 Cytoplasm 0,001417 1,99612969 B2RPK0 Putative high mobility group protein B1-like 1 0,032314 1,590743352 O00231 26S proteasome non-ATPase regulatory subunit 11 Protein metabolism Cytoplasm; Nucleus 0,000029 1,428664042 O00232 26S proteasome non-ATPase regulatory subunit 12 Protein metabolism Cytoplasm 0,008566 1,544545922 O00264 Membrane-associated progesterone receptor component 1 Cell communication Plasma membrane 0,001459 2,322924147 O00429 Dynamin-1-like protein Mitochondrion organization and biogenesis Cytoplasm 0,006560 0,06391487 O00567 Nucleolar protein 56 Regulation of nucleotide metabolism Nucleus 0,007330 3,842896338 O14737 Programmed cell death protein 5 Apoptosis Cytoplasm; Nucleus 0,006358 3,836727237 O14818 Proteasome subunit alpha type-7 Protein metabolism Cytoplasm 0,030521 1,893928387 O14979 Heterogeneous nuclear ribonucleoprotein D-like Regulation of nucleotide metabolism Nucleus 0,000637 2,150005885 O15078 Centrosomal protein of 290 kDa 0,015359 14,26648619 O15347 High mobility group protein B3 Regulation of nucleotide metabolism Nucleus 0,005500 1,364309014 O15540 Fatty acid-binding protein_ brain Transport Cytoplasm 0,000087 3,125786118 O43237 Cytoplasmic dynein 1 light intermediate