Divergent Evolution and Evolution by the Birth-And-Death Process in the Immunoglobulin VH Gene Family
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evolution Activity, Grade 11
Evolution Activity, Grade 11 Evolution Activities for Grade 11 Students at the Toronto Zoo 1 Evolution Activity, Grade 11 Table of Contents Pre-Zoo Activity 3-8 • Think, Pair, Share – Animals in Society and Role of Zoos 3-5 o Description 3 o Materials 3 o Four Corners Activity 6 o Background Information 7-8 Zoo Activity 9-19 • Teacher’s Notes 9-13 o General Information, Curriculum expectations, 9-10 materials, procedure o Evaluation Rubrics 11-12 o Glossary 13 • Student Assignment 14-19 o Part 1 – Mission Preparation at the Zoo 15-16 (Observation Sheets) o Part 2 – Scientific Notes 17 o Part 3 – Documentation: The Story 18 o Appendix – Animal signs 19 Evaluation 20 2 Evolution Activity, Grade 11 Suggested Pre-zoo activity Time needed : 35 minutes (or more) Type of activity : pair-share, small-group (approximately 2-3 students) Objective : encourage students to think about and evaluate the roles of animals in our society and the purposes of zoos along with their own attitudes or stands toward zoos Materials needed : a set of 8-16 statements and a mode of ranking (either above the line-below the line or diamond style ranking system) Special note : In order to manage time, teacher can chose to use any number of the statements as long as the 4 core statements listed bellow are included. Task : students work together to rank the statements about the treatment of animals. They should work together and try to negotiate a consensus, but if this is impossible they can either leave out the particular statement or write down a few lines in their notes as to why they would place them in a different category. -

Divergent Evolution in Antiherbivore Defences Within Species Complexes at a Single Amazonian Site
Journal of Ecology 2015, 103, 1107–1118 doi: 10.1111/1365-2745.12431 Divergent evolution in antiherbivore defences within species complexes at a single Amazonian site Marıa-Jose Endara1*, Alexander Weinhold1, James E. Cox2, Natasha L. Wiggins1, Phyllis D. Coley1,3 and Thomas A. Kursar1,3 1Department of Biology, The University of Utah, Salt Lake City, UT 84112-0840, USA; 2Health Sciences Center Core Research Facilities, School of Medicine, The University of Utah, Salt Lake City, UT 84112-0840, USA; and 3Smithsonian Tropical Research Institute, Balboa Box 0843-03092, Panama Summary 1. Classic theory in plant–insect interactions has linked herbivore pressure with diversification in plant species. We hypothesize that herbivores may exert divergent selection on defences, such that closely related plant species will be more different in defensive than in non-defensive traits. 2. We evaluated this hypothesis by investigating two clades of closely related plant species coexis- ting at a single site in the Peruvian Amazon: Inga capitata Desv. and Inga heterophylla Willd. spe- cies complexes. We compared how these lineages differ in the suite of chemical, biotic, phenological and developmental defences as compared to non-defensive traits that are related to habitat use and resource acquisition. We also collected insect herbivores feeding on the plants. 3. Our data show that sister lineages within both species complexes are more divergent in antiherbi- vore defences than in other non-defensive, functional traits. Moreover, the assemblages of herbivore communities are dissimilar between the populations of coexisting I. capitata lineages. 4. Synthesis. Our results are consistent with the idea that for the I. -

The Genetic Causes of Convergent Evolution
Nature Reviews Genetics | AOP, published online 9 October 2013; doi:10.1038/nrg3483 REVIEWS The genetic causes of convergent evolution David L. Stern Abstract | The evolution of phenotypic similarities between species, known as convergence, illustrates that populations can respond predictably to ecological challenges. Convergence often results from similar genetic changes, which can emerge in two ways: the evolution of similar or identical mutations in independent lineages, which is termed parallel evolution; and the evolution in independent lineages of alleles that are shared among populations, which I call collateral genetic evolution. Evidence for parallel and collateral evolution has been found in many taxa, and an emerging hypothesis is that they result from the fact that mutations in some genetic targets minimize pleiotropic effects while simultaneously maximizing adaptation. If this proves correct, then the molecular changes underlying adaptation might be more predictable than has been appreciated previously. (FIG. 1) Fitness Different species often evolve similar solutions to envi introgression . It is worth distinguishing between The potential evolutionary ronmental challenges. Insects, birds and bats evo these scenarios because each provides evidence for a dif success of a genotype, defined lved wings, and octopi, vertebrates and spiders ferent evolutionary path3. The first case, the independent as the reproductive success or evolved focusing eyes. Phenotypic convergence provides origin and spread of mutations, has been called parallel the proportion of genes that an individual leaves in the gene compelling evidence that ecological circumstances can genetic evolution. I suggest that the evolution of alleles 1,2 pool of the next generation in a select for similar evolutionary solutions . -

Statement About Pnas Paper “The New Mutation Theory of Phenotypic Evolution”
July 30, 2007 (revised) Brief Historical Perspectives of the Paper “THE NEW MUTATION THEORY OF PHENOTYPIC EVOLUTION” Proceedings of the National Academy of Sciences, USA, 104:12235-12242, Published July 17, 2007 online. Masatoshi Nei The following is the author’s account of the history of development of the mutation theory of phenotypic evolution and related matters. Darwinism (1) Charles Darwin proposed that evolution occurs by natural selection in the presence of fluctuating variation. (2) At his time, the mechanism of generation of new variation was not known, and he assumed that new variation is generated by climatic changes, inheritance of acquired characters, spontaneous variations, etc. However, he believed that new variation occurs in random direction and evolutionary change is caused by natural selection. (3) He assumed that enormous phenotypic diversity among different phyla and classes is the result of accumulation of gradual evolution by natural selection. Saltationism Several biologists such as Thomas Huxley, William Bateson, and Hugo de Vries argued that the extent of variation between species is much greater than that within 1 species and therefore saltational or macromutational change is necessary to form new species rather than gradual evolution as proposed by Darwin. In this view a new species can be produced by a single macromutation, and therefore natural selection is unimportant. This view was strengthened when de Vries discovered several new forms of evening primroses which were very different from the parental species (Oenothera lamarckiana) and appeared to be new species. This discovery suggested that new species can arise by a single step of macromutation. However, it was later shown that O. -

Evolution by the Birth-And-Death Process in Multigene Families of the Vertebrate Immune System
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 7799–7806, July 1997 Colloquium Paper This paper was presented at a colloquium entitled ‘‘Genetics and the Origin of Species,’’ organized by Francisco J. Ayala (Co-chair) and Walter M. Fitch (Co-chair), held January 30–February 1, 1997, at the National Academy of Sciences Beckman Center in Irvine, CA. Evolution by the birth-and-death process in multigene families of the vertebrate immune system MASATOSHI NEI*, XUN GU, AND TATYANA SITNIKOVA Institute of Molecular Evolutionary Genetics and Department of Biology, The Pennsylvania State University, 328 Mueller Laboratory, University Park, PA 16802 ABSTRACT Concerted evolution is often invoked to explain the diversity and evolution of the multigene families of major histocompatibility complex (MHC) genes and immunoglobulin (Ig) genes. However, this hypothesis has been controversial because the member genes of these families from the same species are not necessarily more closely related to one another than to the genes from different species. To resolve this controversy, we conducted phylogenetic analyses of several multigene families of the MHC and Ig systems. The results show that the evolutionary pattern of these families is quite different from that of concerted evolution but is in agreement with the birth-and-death model of evolution in which new genes are created by repeated gene duplication and some duplicate genes are maintained in the genome for a long time but others are deleted or become nonfunctional by deleterious mutations. We found little evidence that interlocus gene conversion plays an important role in the evolution of MHC and Ig multigene families. -

The Evolution of Mammalian Aging
Experimental Gerontology 37 &2002) 769±775 www.elsevier.com/locate/expgero The evolution of mammalian aging JoaÄo Pedro de MagalhaÄes*, Olivier Toussaint Department of Biology, Unit of Cellular Biochemistry and Biology, University of Namur FUNDP), Rue de Bruxelles 61, 5000 Namur, Belgium Received 18 December 2001; received in revised form 15 January 2002; accepted 18 January 2002 Abstract The incidence of aging is different between mammals and their closer ancestors &e.g. reptiles and amphibians). While all studied mammals express a well-de®ned aging phenotype, many amphibians and reptiles fail to show signs of aging. In addition, mammalian species show great similarities in their aging phenotype, suggesting that a common origin might be at work. The proposed hypothesis is that mammalian aging evolved together with the ancestry of modern mammals. In turn, this suggests that the fundamental cause of human aging is common to most, if not all, mammals and might be a unique phenom- enon. Experimental procedures capable of testing these theories and how to map the causes of mammalian and thus, human aging, are predicted. q 2002 Elsevier Science Inc. All rights reserved. Keywords: Aging; Senescence; Evolution; Mammals; Reptiles; Birds; Longevity 1. Introduction life span to Marion's tortoise capable of living over a century in captivity. Studies conducted in frogs failed Aging affects all studied mammalian species: from to indicate any increase in mortality both in the wild mice living no more than a mere half-a-dozen years to &Plytycz et al., 1995) and in captivity &Brocas and humans capable of living over 120years &Comfort, VerzaÁr, 1961). -

What Is Natural Selection?
This week (June 8th – 12th) we are reviewing evolution. 1. Review the lecture on evolution 2. Watch the evidence of evolution video: https://www.khanacademy.org/science/high-school-biology/hs-evolution/hs- evidence-of-evolution/v/evidence-for-evolution 3. Watch the Types of Natural Selection video https://youtu.be/64JUJdZdDQo 4. Complete the Evolution Review Activity 5. Complete the Natural Selection Reading and Questions I am grading you based on the rubric below. If you get below a 70% I will allow you to redo the assignment. Evidence of Evolution & the Week 5 Remote Learning Mechanisms of Biodiversity Natural Selection The total variety of all the organisms in the biosphere= Biodiversity Where did all these different organisms come from? How are they related? Some organisms in a population are less likely to survive. Over time, natural selection results in changes in the inherited characteristics of a population. WHAT IS DARWIN’S THEORY? These changes increase a species’ fitness in its environment. Any inherited characteristic Ability of an individual to that increases an organism’s survive and reproduce in its chance of survival= specific environment= FITNESS ADAPTATION WHAT IS DARWIN’S THEORY? STRUGGLE FOR EXISTENCE: means that members of each species must compete for food, space, and other resources. SURVIVAL OF THE FITTEST: organisms which are better adapted to the environment will survive and reproduce, passing on their genes. DESCENT WITH MODIFICATION: suggests that each species has decended, with changes, from other species over time. • This idea suggests that all living species are related to each other, and that all species, living and extinct,share a common ancestor. -

Evolution of Ant-Mimicking Beetles Edcator Materials
Evolution of Ant-Mimicking Beetles Data Point Educator Materials HOW TO USE THIS RESOURCE Show the following figures and caption to your students. The accompanying Student Handout provides space below the caption for Observations, Notes, and Questions and space next to the “Background Information” for Big Ideas, Notes, and Questions. The “Interpreting the Figures” and “Discussion Questions” sections provide additional information and suggested questions that you can use to prompt student thinking, increase engagement, or guide a class discussion about the characteristics of the figures and what they show. Figure 1: Figure 2: Caption: Figure 1 shows one species of ant-mimicking rove beetle (front) next to the ant species (back) that it mimics. The top half of Figure 2 shows more species of rove beetles from the same subfamily, Aleocharinae. “Free- living” beetles (example shown on the left) have a generalized beetle body type. “Army ant social parasite” beetles (examples shown on the right) have a specialized ant-mimicking (also called “myrmecoid”) body type. The bottom half of Figure 2 shows the evolutionary relationships among multiple Aleocharinae lineages that are either generalized (black) or ant-mimicking (orange). The labeled circle indicates the most recent common ancestor of all the ant-mimicking lineages. Phylogeny Published November 2019 www.BioInteractive.org Page 1 of 3 Data Point Evolution of Ant-Mimicking Beetles Educator Materials BACKGROUND INFORMATION Rove beetles are a large family of beetle species found all over the world. Although most rove beetles live freely on their own, many species in the subfamily Aleocharinae live closely with another type of insect: army ants. -

Book Reviews
Heredity 86 (2001) 385±386 Book reviews Molecular Evolution and Phylogenetics. Masatoshi Nei and will ®nd Molecular Evolution and Phylogenetics as useful as its Sudhir Kumar. Oxford University Press, Oxford. 2000. previous incarnation and all readers will come away with the Pp. 333. Price £65.00, hardback. ISBN 0 19 513584 9. fully justi®ed feeling that there is a lot more to this tree building business than ®rst meets the eye. We live in interesting times. The curse is that it is near impossible to keep up with a ®eld moving as rapidly as molecular evolution. The blessing is that we have clear- References writing proponents, such as the authors of this book, to help FELSENSTEIN, J. 2001. Inferring Phylogenies. Sinauer Associates, Inc., us with comprehensive and comprehensible reviews. Molecu- Sunderland, Massachusetts. lar Evolution and Phylogenetics is essentially a major update LI, W.-H. 1997. Molecular Evolution. Sinauer Associates, Inc., Sunder- of Nei's 1987 book, Molecular Evolutionary Genetics. The land, Massachusetts. title of the newer volume from the doyen of distance methods NEI, M. 1987. Molecular Evolutionary Genetics. Columbia University accurately re¯ects its much greater emphasis on phylogenetic Press, New York. issues. The book begins with chapters on the molecular basis PAGE, R. D. M.AND HOLMES , E. C. 1998 Molecular Evolution: A of evolution, evolutionary change in amino acid and DNA Phylogenetic Approach. Blackwell Science Ltd., Oxford. sequences, and models of nucleotide substitutions. Inferring SWOFFORD, D. L., OLSEN, G. J. , WADDELLP. J.AND HILLIS , D. M. 1996. phylogenies from molecular sequence data is a fast moving Phylogenetic inference. -

Molecular Evolutionary Genetics. by Masatoshi Nei. New York
Book reviews 74 Barry Polisky discusses replication control of recommendation, only mitigated by the high price, ColEl-type plasmids and Robert Knowlton considers which will limit its siting to library rather than copy number and stability of yeast plasmids, in the personal bookshelves. next two chapters; and in both systems the implica- ERIC REEVE tions for maximising expression of cloned genes are Institute of Animal Genetics examined. These form important current areas of University of Edinburgh applied research and the authors examine the problems which each system presents. Gary Stormo discusses translation initiation in both prokaryotes and eukaryotes. Some laboratories have Molecular Evolutionary Genetics. By MASATOSHI NEI. begun to collect data on the efficiencies of particular New York: Columbia University Press. 1987. 512 initiation sites, and Stormo concentrates on the effects pages. U.S. $50.00. ISBN 0 231 06320 2. of mRNA sequence on translation efficiency. This, of The rapid accumulation of data at the molecular course, brings up the very difficult problem of genetic level, from protein and DNA sequencing, structural features of the mRNA, which are difficult restriction enzyme analysis and electrophoretic vari- to predict from its sequence but can have an important ants have given us much information on the tempo effect on initiation rate. The E. coli data lead to the and structure of the evolutionary process and on following rules for maximizing the yield of a particular variability within populations. Major theoretical protein: (1) AUG is the best initiation codon; (2) A developments have ensued in order to interpret all the Shine-Dalgardo sequence of at least four nucleotides, new data. -

Evolutionary Origins for Ecological Patterns in Space PERSPECTIVE Mark C
PERSPECTIVE Evolutionary origins for ecological patterns in space PERSPECTIVE Mark C. Urbana,b,1, Sharon Y. Straussc, Fanie Pelletierd, Eric P. Palkovacse, Mathew A. Leiboldf, Andrew P. Hendryg,h, Luc De Meesteri,j,k, Stephanie M. Carlsonl, Amy L. Angertm, and Sean T. Gierya,n Edited by Simon A. Levin, Princeton University, Princeton, NJ, and approved June 5, 2020 (received for review January 24, 2020) Historically, many biologists assumed that evolution and ecology acted independently because evolution occurred over distances too great to influence most ecological patterns. Today, evidence indicates that evolution can operate over a range of spatial scales, including fine spatial scales. Thus, evolutionary divergence across space might frequently interact with the mechanisms that also determine spatial ecological patterns. Here, we synthesize insights from 500 eco-evolutionary studies and develop a predictive framework that seeks to understand whether and when evolution amplifies, dampens, or creates ecological patterns. We demonstrate that local adaptation can alter everything from spatial variation in population abundances to ecosystem properties. We uncover 14 mechanisms that can mediate the outcome of evolution on spatial ecological patterns. Sometimes, evolution amplifies environmental variation, especially when selection enhances resource uptake or patch selection. The local evolution of foundation or keystone species can create ecological patterns where none existed originally. However, most often, we find that evolution dampens existing environmental gradients, because local adaptation evens out fitness across environments and thus counteracts the variation in associated ecological patterns. Consequently, evolution generally smooths out the underlying heterogeneity in nature, making the world appear less ragged than it would be in the absence of evolution. -
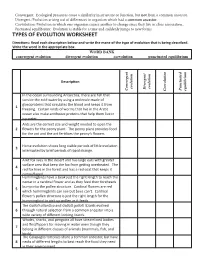
TYPES of EVOLUTION WORKSHEET Directions: Read Each Description Below and Write the Mane of the Type of Evolution That Is Being Described
Convergent: Ecological pressures cause a similarity in structure or function, but not from a common ancestor. Divergent: Evolution arising out of differences in organism which had a common ancestor. Coevolution: Evolution in which one organism causes another to change since they live in close association.. Pnctuated equilibrium: Evolution is stable for a time and suddenly jumps to new forms TYPES OF EVOLUTION WORKSHEET Directions: Read each description below and write the mane of the type of evolution that is being described. Write the word in the appropriate box. WORD BANK convergent evolution divergent evolution coevolution punctuated equilibrium Description evolution evolution Divergent Punctuated Convergent equilibrium Coevolution In the ocean surrounding Antarctica, there are fish that survive the cold water by using a molecule made of glycoproteins that circulates the blood and keeps it from 1 freezing. Certain kinds of worms that live in the Arctic ocean also make antifreeze proteins that help them live in icy water. Ants are the correct size and weight needed to open the 2 flowers for the peony plant. The peony plant provides food for the ant and the ant fertilizes the peony’s flowers Horse evolution shows long stable periods of little evolution 3 interrupted by brief periods of rapid change. A kit fox lives in the desert and has large ears with greater surface area that keep the fox from getting overheated. The 4 red fox lives in the forest and has a red coat that keeps it camouflaged. Hummingbirds have a beak just the right length to reach the nectar in a cardinal flower and as they feed their foreheads bump into the pollen structure.