Independent Apoptotic Pathways
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Greg's Awesome Thesis
Analysis of alignment error and sitewise constraint in mammalian comparative genomics Gregory Jordan European Bioinformatics Institute University of Cambridge A dissertation submitted for the degree of Doctor of Philosophy November 30, 2011 To my parents, who kept us thinking and playing This dissertation is the result of my own work and includes nothing which is the out- come of work done in collaboration except where specifically indicated in the text and acknowledgements. This dissertation is not substantially the same as any I have submitted for a degree, diploma or other qualification at any other university, and no part has already been, or is currently being submitted for any degree, diploma or other qualification. This dissertation does not exceed the specified length limit of 60,000 words as defined by the Biology Degree Committee. November 30, 2011 Gregory Jordan ii Analysis of alignment error and sitewise constraint in mammalian comparative genomics Summary Gregory Jordan November 30, 2011 Darwin College Insight into the evolution of protein-coding genes can be gained from the use of phylogenetic codon models. Recently sequenced mammalian genomes and powerful analysis methods developed over the past decade provide the potential to globally measure the impact of natural selection on pro- tein sequences at a fine scale. The detection of positive selection in particular is of great interest, with relevance to the study of host-parasite conflicts, immune system evolution and adaptive dif- ferences between species. This thesis examines the performance of methods for detecting positive selection first with a series of simulation experiments, and then with two empirical studies in mammals and primates. -

Network Pharmacology Interpretation of Fuzheng–Jiedu Decoction Against Colorectal Cancer
Hindawi Evidence-Based Complementary and Alternative Medicine Volume 2021, Article ID 4652492, 16 pages https://doi.org/10.1155/2021/4652492 Research Article Network Pharmacology Interpretation of Fuzheng–Jiedu Decoction against Colorectal Cancer Hongshuo Shi ,1 Sisheng Tian,2 and Hu Tian 3 1College of Traditional Chinese Medicine, Shandong University of Traditional Chinese Medicine, Jinan, Shandong, China 2School of Management, Shandong University of Traditional Chinese Medicine, Jinan, Shandong, China 3College of Traditional Chinese Medicine, Shandong University of Traditional Chinese Medicine, Jinan, Shandong, China Correspondence should be addressed to Hu Tian; [email protected] Received 7 April 2020; Revised 3 January 2021; Accepted 21 January 2021; Published 20 February 2021 Academic Editor: George B. Lenon Copyright © 2021 Hongshuo Shi et al. ,is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Introduction. Traditional Chinese medicine (TCM) believes that the pathogenic factors of colorectal cancer (CRC) are “deficiency, dampness, stasis, and toxin,” and Fuzheng–Jiedu Decoction (FJD) can resist these factors. In this study, we want to find out the potential targets and pathways of FJD in the treatment of CRC and also explain from a scientific point of view that FJD multidrug combination can resist “deficiency, dampness, stasis, and toxin.” Methods. We get the composition of FJD from the TCMSP database and get its potential target. We also get the potential target of colorectal cancer according to the OMIM Database, TTD Database, GeneCards Database, CTD Database, DrugBank Database, and DisGeNET Database. -

Genetic Variation As a Tool for Identifying Novel Transducers of Itch
Genetic variation as a tool for identifying novel transducers of itch By Takeshi Morita A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Molecular and Cell Biology in the Graduate Division of the University of California, Berkeley Committee in charge: Professor Diana M. Bautista, Co-Chair Professor Rachel B. Brem, Co-Chair Professor John Ngai Professor Kristin Scott Professor Michael W. Nachman Summer 2016 Abstract Genetic variation as a tool for identifying novel transducers of itch by Takeshi Morita Doctor of Philosophy in Molecular and Cell Biology University of California, Berkeley Professor Diana M. Bautista, Co-Chair Professor Rachel B. Brem, Co-Chair The mammalian somatosensory system mediates itch, the irritating sensation that elicits a desire to scratch. Millions of people worldwide suffer from chronic itch that fails to respond to current drugs and therapies. Even though recent studies have begun to elucidate the basic characteristics of the itch circuitry, we have little understanding about the molecules and signaling mechanisms that underlie detection and transduction of itch sensation, especially during chronic itch conditions. We have taken a genomic approach by harnessing natural variation in itch-evoked scratching behaviors in mice to identify novel molecular players that are involved in itch signal transduction at the level of primary sensory neurons. From our analysis, we identified numerous candidate itch genes, and further identified a serotonin receptor, HTR7 as a key transducer that is required for both development and maintenance of chronic itch. We further investigated the genetic basis of variation in itch, and identified a set of genes and regulatory pathways that may be involved in controlling itch behaviors. -

Exploring the Pharmacological Mechanism of Quercetin-Resveratrol Combination for Polycystic Ovary Syndrome
www.nature.com/scientificreports Corrected: Publisher Correction OPEN Exploring the Pharmacological Mechanism of Quercetin- Resveratrol Combination for Polycystic Ovary Syndrome: A Systematic Pharmacological Strategy-Based Research Kailin Yang1,2,6, Liuting Zeng 3,6*, Tingting Bao3,4,6, Zhiyong Long5 & Bing Jin3* Resveratrol and quercetin have efects on polycystic ovary syndrome (PCOS). Hence, resveratrol combined with quercetin may have better efects on it. However, because of the limitations in animal and human experiments, the pharmacological and molecular mechanism of quercetin-resveratrol combination (QRC) remains to be clarifed. In this research, a systematic pharmacological approach comprising multiple compound target collection, multiple potential target prediction, and network analysis was used for comparing the characteristic of resveratrol, quercetin and QRC, and exploring the mechanism of QRC. After that, four networks were constructed and analyzed: (1) compound-compound target network; (2) compound-potential target network; (3) QRC-PCOS PPI network; (4) QRC-PCOS- other human proteins (protein-protein interaction) PPI network. Through GO and pathway enrichment analysis, it can be found that three compounds focus on diferent biological processes and pathways; and it seems that QRC combines the characteristics of resveratrol and quercetin. The in-depth study of QRC further showed more PCOS-related biological processes and pathways. Hence, this research not only ofers clues to the researcher who is interested in comparing the diferences among resveratrol, quercetin and QRC, but also provides hints for the researcher who wants to explore QRC’s various synergies and its pharmacological and molecular mechanism. Polycystic ovary syndrome (PCOS) is one of the most common female endocrine diseases characterized by hyperandrogenism, menstrual disorders and infertility. -

Structural and Biochemical Characterization of the Human Cyclophilin Family of Peptidyl-Prolyl Isomerases
Structural and Biochemical Characterization of the Human Cyclophilin Family of Peptidyl-Prolyl Isomerases Tara L. Davis1,2¤, John R. Walker1, Vale´rie Campagna-Slater1, Patrick J. Finerty, Jr.1, Ragika Paramanathan1, Galina Bernstein1, Farrell MacKenzie1, Wolfram Tempel1, Hui Ouyang1, Wen Hwa Lee1,3, Elan Z. Eisenmesser4, Sirano Dhe-Paganon1,2* 1 Structural Genomics Consortium, University of Toronto, Toronto, Ontario, Canada, 2 Department of Physiology, University of Toronto, Toronto, Ontario, Canada, 3 University of Oxford, Headington, United Kingdom, 4 Department of Biochemistry & Molecular Genetics, University of Colorado Denver, Aurora, Colorado, United States of America Abstract Peptidyl-prolyl isomerases catalyze the conversion between cis and trans isomers of proline. The cyclophilin family of peptidyl-prolyl isomerases is well known for being the target of the immunosuppressive drug cyclosporin, used to combat organ transplant rejection. There is great interest in both the substrate specificity of these enzymes and the design of isoform-selective ligands for them. However, the dearth of available data for individual family members inhibits attempts to design drug specificity; additionally, in order to define physiological functions for the cyclophilins, definitive isoform characterization is required. In the current study, enzymatic activity was assayed for 15 of the 17 human cyclophilin isomerase domains, and binding to the cyclosporin scaffold was tested. In order to rationalize the observed isoform diversity, the high-resolution crystallographic structures of seven cyclophilin domains were determined. These models, combined with seven previously solved cyclophilin isoforms, provide the basis for a family-wide structure:function analysis. Detailed structural analysis of the human cyclophilin isomerase explains why cyclophilin activity against short peptides is correlated with an ability to ligate cyclosporin and why certain isoforms are not competent for either activity. -

Dissection of Xist Functional Elements Involved in X-Chromosome Inactivation
Dissection of Xist Functional Elements Involved in X-Chromosome Inactivation The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Colognori, David A. 2019. Dissection of Xist Functional Elements Involved in X-Chromosome Inactivation. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:42029476 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Dissection of Xist functional elements involved in X-chromosome inactivation A dissertation presented by David A. Colognori to The Division of Medical Sciences in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the subject of Biological and Biomedical Sciences Harvard University Cambridge, Massachusetts May 2019 © 2019 by David A. Colognori All rights reserved. Dissertation Advisor: Jeannie T. Lee David A. Colognori Dissection of Xist functional elements involved in X-chromosome inactivation Abstract X-chromosome inactivation (XCI) is the epigenetic process of silencing one of the two X chromosomes in female mammals to balance X-linked gene dosage with that of males. This process is governed by the long noncoding RNA Xist. Xist is expressed from and coats one X chromosome in cis, leading to recruitment of various protein factors to silence gene expression. However, the functional RNA elements and molecular mechanisms involved in Xist coating and silencing remain ambiguous. -

(12) United States Patent (10) Patent No.: US 7,199,281 B2 Murray Et Al
US007 199281B2 (12) United States Patent (10) Patent No.: US 7,199,281 B2 Murray et al. (45) Date of Patent: Apr. 3, 2007 (54) METHOD OF GENERATING ATRANSGENIC Seamark, 1994, Reproductive Fertility and Development, 6:653-7.* LIVESTOCK ANIMAL Mullins, 1996, J. Clin. Invest., vol. 98, pp. S37-S40.* McCreath, 2000, Nature, vol. 405, pp. 1066-1069.* (75) Inventors: James D. Murray, Davis, CA (US); Denning, C. 2001 A. Nature Biotechnology, vol. 19, pp. 559-562.* Elizabeth A. Maga, Sacramento, CA Dinnyes, 2002, Cloning and Stem Cells, vol. 4, pp. 81-90.* (US); Gary B. Anderson, Davis, CA Denning, C., 2001B, Gene Targeting from primaty fetal fibroblasts (US); Stefanie M. Oppenheim, Davis, from sheep and pig, Cloning and Stem Cells, 3:221-231.* CA (US) Poljaeva, I.A., 2000, Nature, 407:86-90.* Clark, A.J., 2000, Gene targeting in livestock: a preview, Transgenic (73) Assignee: The Regents of the University of Research, 9:263-275.* California, Oakland, CA (US) Capecchi, 1994, Targeted Gene Replacement, Scientific American, vol. 270, pp. 34-41.* (*) Notice: Subject to any disclaimer, the term of this http://en.wikipedia.org/wiki/Livestock.* patent is extended or adjusted under 35 Harrison, SJ et al., 2002, Efficient generation of 1(1.3) U.S.C. 154(b) by 412 days. galactosyltransferase knockout porcine fetal fibroblasts for nuclear transfer, Transgenic Research, 11:143-150.* (21) Appl. No.: 10/238,042 Poljaeva I.A. and Campbell, KHS, 2000, New advances in somatic cell nuclear transfer:application in transgenesis, Theriogenology, (22) Filed: Sep. 6, 2002 53:117-126. Thomson, AJ et al., 2003, Gene targeting in livestock, Reproduction (65) Prior Publication Data Supplement, 61:495-508.* US 2003/O115618 A1 Jun. -

System-Wide Identification of Wild-Type SUMO-2 Conjugation Sites
ARTICLE Received 16 Oct 2014 | Accepted 26 Apr 2015 | Published 15 Jun 2015 DOI: 10.1038/ncomms8289 OPEN System-wide identification of wild-type SUMO-2 conjugation sites Ivo A. Hendriks1, Rochelle C. D’Souza2, Jer-Gung Chang1, Matthias Mann2 & Alfred C.O. Vertegaal1 SUMOylation is a reversible post-translational modification (PTM) regulating all nuclear processes. Identification of SUMOylation sites by mass spectrometry (MS) has been hampered by bulky tryptic fragments, which thus far necessitated the use of mutated SUMO. Here we present a SUMO-specific protease-based methodology which circumvents this problem, dubbed Protease-Reliant Identification of SUMO Modification (PRISM). PRISM allows for detection of SUMOylated proteins as well as identification of specific sites of SUMOylation while using wild-type SUMO. The method is generic and could be widely applied to study lysine PTMs. We employ PRISM in combination with high-resolution MS to identify SUMOylation sites from HeLa cells under standard growth conditions and in response to heat shock. We identified 751 wild-type SUMOylation sites on endogenous proteins, including 200 dynamic SUMO sites in response to heat shock. Thus, we have developed a method capable of quantitatively studying wild-type mammalian SUMO at the site-specific and system-wide level. 1 Department of Molecular Cell Biology, Leiden University Medical Center, Albinusdreef 2, 2333 ZA Leiden, the Netherlands. 2 Department for Proteomics and Signal Transduction, Max Planck Institute for Biochemistry, Am Klopferspitz 18, D-82152 Martinsried, Germany. Correspondence and requests for materials should be addressed to A.C.O.V. (email: [email protected]). NATURE COMMUNICATIONS | 6:7289 | DOI: 10.1038/ncomms8289 | www.nature.com/naturecommunications 1 & 2015 Macmillan Publishers Limited. -

HHS Public Access Provided by Iupuischolarworks Author Manuscript
View metadata, citation and similar papers at core.ac.uk brought to you by CORE HHS Public Access provided by IUPUIScholarWorks Author manuscript Author Manuscript Author ManuscriptPharmacol Author Manuscript Biochem Behav Author Manuscript . Author manuscript; available in PMC 2015 July 27. Published in final edited form as: Pharmacol Biochem Behav. 2008 June ; 89(4): 481–498. doi:10.1016/j.pbb.2008.01.023. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats Zachary A. Rodd1,6, Mark W. Kimpel1,6, Howard J. Edenberg2,4,5, Richard L. Bell1,6, Wendy N. Strother1,6, Jeanette N. McClintick2,5, Lucinda G. Carr3, Tiebing Liang3, and William J. McBride1,6 1Department of Psychiatry, Indiana University School of Medicine, Indianapolis, IN 46202-4887 2Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, IN 46202-4887 3Department of Medicine, Indiana University School of Medicine, Indianapolis, IN 46202-4887 4Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN 46202-4887 5Center for Medical Genomics, Indiana University School of Medicine, Indianapolis, IN 46202-4887 6Institute of Psychiatric Research, Indiana University School of Medicine, Indianapolis, IN 46202-4887 Abstract The current study examined the effects of operant ethanol (EtOH) self-administration on gene expression in the nucleus accumbens (ACB) and amygdala (AMYG) of inbred alcohol-preferring (iP) rats. Rats self-trained on a standard two-lever operant paradigm to administer either water- water, EtOH (15% v/v)-water, or saccharin (SAC; 0.0125% g/v)-water. Animals were killed 24 hr after the last operant session, and the ACB and AMYG dissected; RNA was extracted and purified for microarray analysis. -

Modifier Genes for Mouse Phosphatidylinositol Transfer Protein
Genetics: Published Articles Ahead of Print, published on February 14, 2011 as 10.1534/genetics.110.125906 Modifier genes for mouse Phosphatidylinositol Transfer Protein alpha (vibrator) that bypass juvenile lethality. Dorothy Concepcion*, Frank Johannes§, Yuan Hung Lo*, Jay Yao*, Jerry Fong*,1 and Bruce A. Hamilton* * Department of Medicine, Department of Cellular & Molecular Medicine, and Institute for Genomic Medicine University of California San Diego 9500 Gilman Drive, La Jolla, CA 92093-0644 § Groningen Bioinformatics Centre University of Groningen 9751NN Haren, The Netherlands 1 Present address: Huffington Center on Aging, 1 Baylor Plaza, Baylor College of Medicine, Houston, TX 77030 [email protected] Copyright 2011. Running title: Modifiers of vibrator lethality Keywords: linkage, QTL, survival analysis, Mvb2, Mvb3 Corresponding Author: Dr. Bruce A. Hamilton George Palade Laboratories for Cellular and Molecular Medicine, Room 256 UCSD School of Medicine 9500 Gilman Drive La Jolla, CA 92093-0644 Telephone: (858) 822-1055 Electronic mail: [email protected] ABSTRACT Phosphatidylinositol transfer proteins (PITPs) mediate lipid signaling and membrane trafficking in eukaryotic cells. Loss-of-function mutations of the gene encoding PITPα in mice result in a range of dosage-sensitive phenotypes, including neurological dysfunction, neurodegeneration and premature death. We have previously reported genetic suppression of a strong hypomorphic allele, vibrator, by a wild-derived variant of Nxf1, which increases the level of PITPα made from vibrator alleles and suppresses each of the neurological and survival phenotypes. Here we report discovery and genetic mapping of additional vibrator modifiers, Mvb2 and Mvb3, from a different strain background that suppresses juvenile lethality without suppressing visible phenotypes or gene expression. -
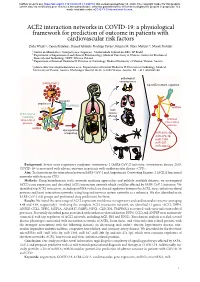
ACE2 Interaction Networks in COVID-19
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.13.094714; this version posted May 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. ACE2 interaction networks in COVID-19: a physiological framework for prediction of outcome in patients with cardiovascular risk factors Zofia Wicik1,2, Ceren Eyileten2, Daniel Jakubik2, Rodrigo Pavão1, Jolanta M. Siller-Matula2,3*, Marek Postula2 ¹ Centro de Matemática, Computação e Cognição - Universidade Federal do ABC, SP, Brazil ² Department of Experimental and Clinical Pharmacology, Medical University of Warsaw, Center for Preclinical Research and Technology CEPT, Warsaw, Poland ³ Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Vienna, Austria * [email protected]; Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Austria. Waehringer Guertel 18-20, A-1090 Vienna, Austria. Tel. +43 1 4040046140 pathological consequences SARS-CoV-2 top ACE2 network regulators activation attachment TMPRSS2 miRNA ACE2 main ACE2 network tissues containing affected virus-related activated proteins virus-related proteins miRNA dysregulation of signaling miR-302c-5p miR-27a-3p miR-1305 miR-587 miR-26b-5p Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (coronavirus disease 2019; COVID-19) is associated with adverse outcome in patients with cardiovascular disease (CVD). Aim: To characterize the interaction between SARS-CoV-2 and Angiotensin Converting Enzyme 2 (ACE2) functional networks with focus on CVD. -
Transcriptome Profiling of Porcine Testis Tissue Reveals Genes Related
Son et al. BMC Veterinary Research (2020) 16:161 https://doi.org/10.1186/s12917-020-02373-9 RESEARCH ARTICLE Open Access Transcriptome profiling of porcine testis tissue reveals genes related to sperm hyperactive motility Maren van Son1* , Nina Hårdnes Tremoen2,3, Ann Helen Gaustad1,2, Dag Inge Våge3, Teklu Tewoldebrhan Zeremichael2, Frøydis Deinboll Myromslien2 and Eli Grindflek1 Abstract Background: Sperm hyperactive motility has previously been shown to influence litter size in pigs, but little is known about the underlying biological mechanisms. The aim of this study was to use RNA sequencing to investigate gene expression differences in testis tissue from Landrace and Duroc boars with high and low levels of sperm hyperactive motility. Boars with divergent phenotypes were selected based on their sperm hyperactivity values at the day of ejaculation (day 0) (contrasts (i) and (ii) for Landrace and Duroc, respectively) and on their change in hyperactivity between day 0 and after 96 h liquid storage at 18 °C (contrast (iii)). Results: RNA sequencing was used to measure gene expression in testis. In Landrace boars, 3219 genes were differentially expressed for contrast (i), whereas 102 genes were differentially expressed for contrast (iii). Forty-one differentially expressed genes were identified in both contrasts, suggesting a functional role of these genes in hyperactivity regardless of storage. Zinc finger DNLZ was the most up-regulated gene in contrasts (i) and (iii), whereas the most significant differentially expressed gene for the two contrasts were ADP ribosylation factor ARFGAP1 and solute carrier SLC40A1, respectively. For Duroc (contrast (ii)), the clustering of boars based on their gene expression data did not reflect their difference in sperm hyperactivity phenotypes.